Applying Genomics to Study and Treat Infectious Diseases
Guest Author: Liz Lucero
There is more to be done to find effective treatments for global health issues like the coronavirus, but genomics can accelerate infectious disease research and vaccine development.
Recently, 10x Genomics CEO Serge Saxonov was asked about the novel coronavirus, 2019-nCoV, and what it will take to address this and other viral threats to worldwide human health.

His answer? Genomics.
While creating a plan for treatment is the ultimate goal, there are several things researchers need to learn about the virus first. In the past, diagnosing, studying, and treating viral outbreaks has been immensely difficult, due in large part to scientific limitations. However, genomic advances have made the process more streamlined than ever. By accessing the genomic sequence of a virus, scientists can interrogate its biology, and use the information gleaned about its structural and functional make-up to answer key questions about its detection, origin, and behavior, and ultimately, find a method for treatment.
Virus Detection
One of the first things scientists need to figure out when facing a potential viral outbreak is detection. They need a quick, reliable way to diagnose the disease in order to track and contain the spread of the virus. Otherwise, it can quickly get out of control. But, diagnosis can’t just rely on symptoms, especially when many viruses can display similar symptoms while varying widely at a molecular level. That’s where genomics comes in. Now, with advances in qPCR, scientists can sequence a virus and use this information to create diagnostic assays that make disease detection relatively simple.
Virus Origins
Knowing where a virus came from can help answer a lot of questions—how and at what rate it spreads, the likelihood of fatality, and how it might mutate, among other things. And to understand a virus’s origins, scientists need to track its entire genome. Discussing 2019-nCoV, Saxonov says, “To understand more deeply the potential origins of the virus, its history and track its evolution, we’ll need to use sequencing technologies (so, not just detect whether it’s present, but determine its full sequence). The sequencing information is crucial to understand how the virus mutates over time as well as across populations.” Researchers have already begun using this method while studying the 2019-nCoV sequence. After comparing the sequences of samples from 10 different patients, scientists observed very little variation, suggesting that the virus has only recently emerged in the human population and has not undergone much mutation yet (1). And using this existing sequencing information, scientists may be able to observe and study further adaptations as they happen.
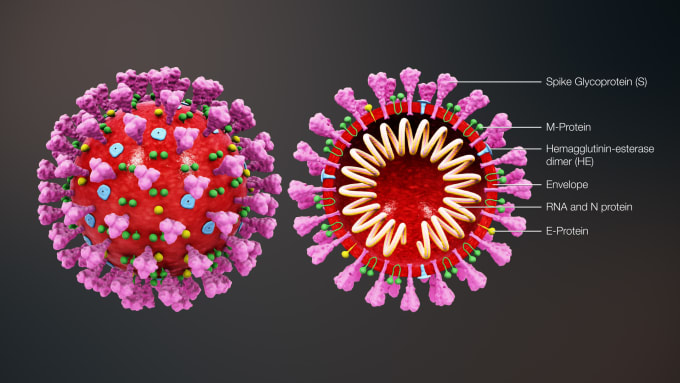
Virus Behavior and Human Response
Another key factor in the study of disease is understanding the biology of the virus. How does the virus behave? How does it infect cells, and how does the human immune system, and more broadly, the body, react and adapt in response? Learning how the human body responds to a virus can provide insight into the incubation period of a virus, how and at what rate it spreads, and why some people can fight off a virus while others cannot. This information can help with containment and, eventually, treatment. But to do this, researchers need to be able to see the whole picture. As Saxonov points out, “it’s crucial to be able to see what is happening in a wide representation of patients’ cells with a high degree of comprehensiveness. We need to do these analyses looking at many genes and gene programs simultaneously because these systems are very complex.” Advances in sequencing, such as single cell RNA-sequencing, have given scientists the ability to access a global view of a patient’s immune response by allowing them to profile a massive number of individual cells. Researchers can now measure, from a mixed sample, the phenotypic or gene expression changes of single cells in response to infection. Such insights can uncover the changes that immune cells undergo as they respond to infected cells, the mechanisms of successful resistance to infection, and even how the virus adapts to these changes.
Accelerating the Path from Discovery to Treatment
With access to genomic information about the adaptive behavior of both the virus and the human immune system, scientists can begin to develop a plan for treatment. Saxonov says, “genomics-based analyses... can be used to find antibodies in patients that can be protective for the broader population. They can also be used to help design vaccines, driven by the understanding of the underlying biology and a precise view of how these vaccines affect different patients.” Indeed, ongoing research continues to validate the use of genomics-based technology for the study of infectious diseases and antibody discovery efforts. Dr. James Crowe of the Vanderbilt University Medical Center, in partnership with the Human Vaccines Project, demonstrated the benefit of single cell technology in his team’s study of antigen-specific B cells and plasmablasts responding to influenza. Additionally, Ian Setliff and colleagues from Vanderbilt University recently published a study in Cell describing their use of single cell immune profiling to rapidly screen thousands of antigen-reactive B cells from two HIV-infected subjects and identify the paired heavy- and light-chain sequence of a novel broadly neutralizing antibody against the virus. With the ability to survey many B cells and full antibody sequences at once, researchers can not only discover new antibodies quickly, but also accelerate production of recombinant antibodies. This can, in turn, be used for prophylactic antibody-based vaccine strategies.
There is much left to be done in the global effort to find effective treatments for the coronavirus. However, there is hope that with global collaboration and access to powerful technology, a successful treatment can be found. In fact, with the wealth of genomic information that has been made available about the coronavirus, researchers have already begun to develop potential vaccines. “There are many scientists across the world working on this challenge. It’s inspiring to see how quickly we can bring so many cutting-edge technologies to bear on this problem. While we are still in the early days of what all of these technologies will enable, it’s amazing to see how much more sophistication we can apply to this challenge compared to just a few years ago,” says Saxonov.
Please contact us if you have more questions about how single cell sequencing workflows can support your study of the coronavirus and other infectious diseases.
References
- Yasinki E, “Scientists Scrutinize New Coronavirus Genome for Answers.”The Scientist.23 Jan 2020. https://www.the-scientist.com/news-opinion/scientist-scrutinize-new-coronavirus-genome-for-answers-67006
