CytAssist Spatial Gene Expression
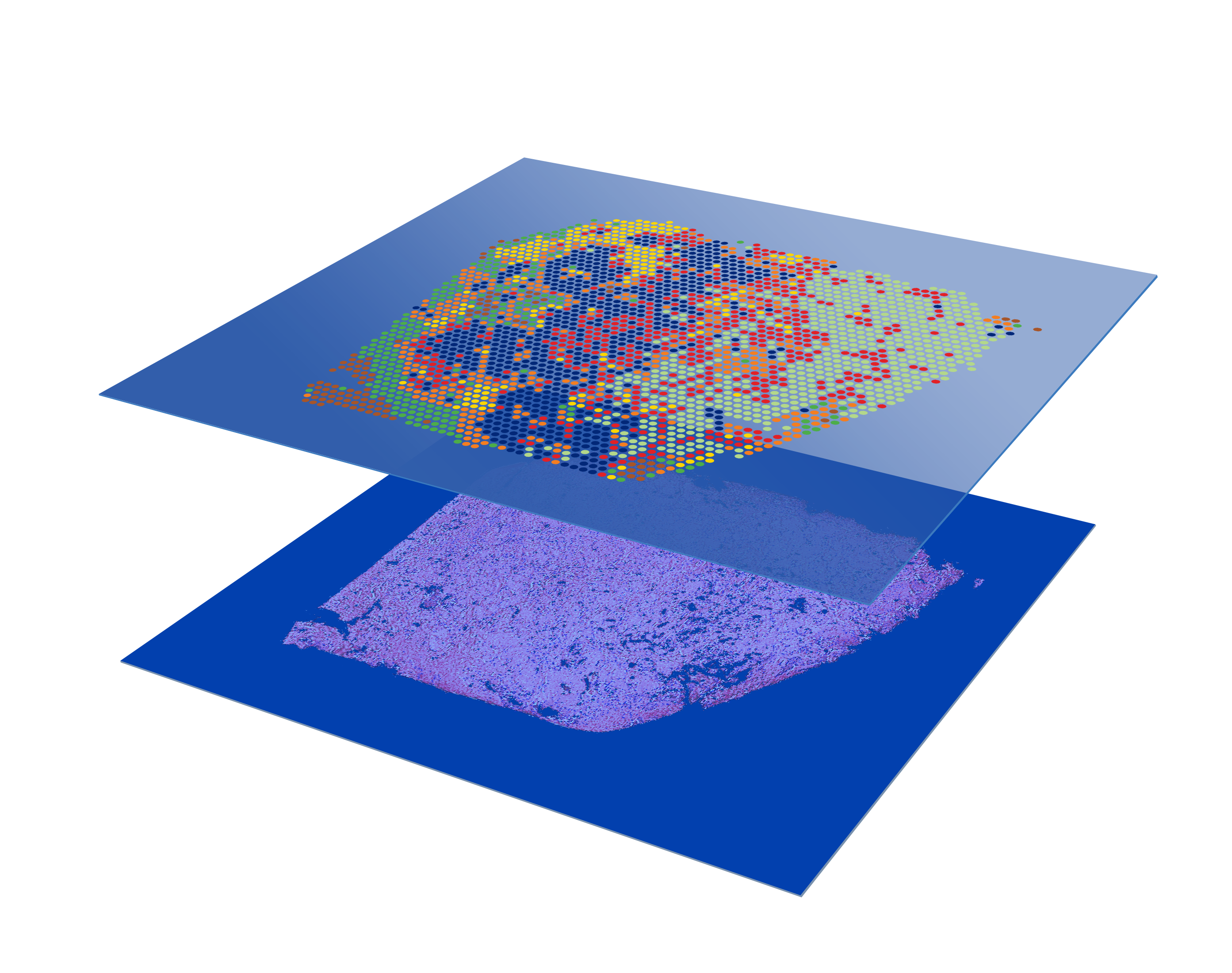
What is CytAssist Spatial Gene Expression?
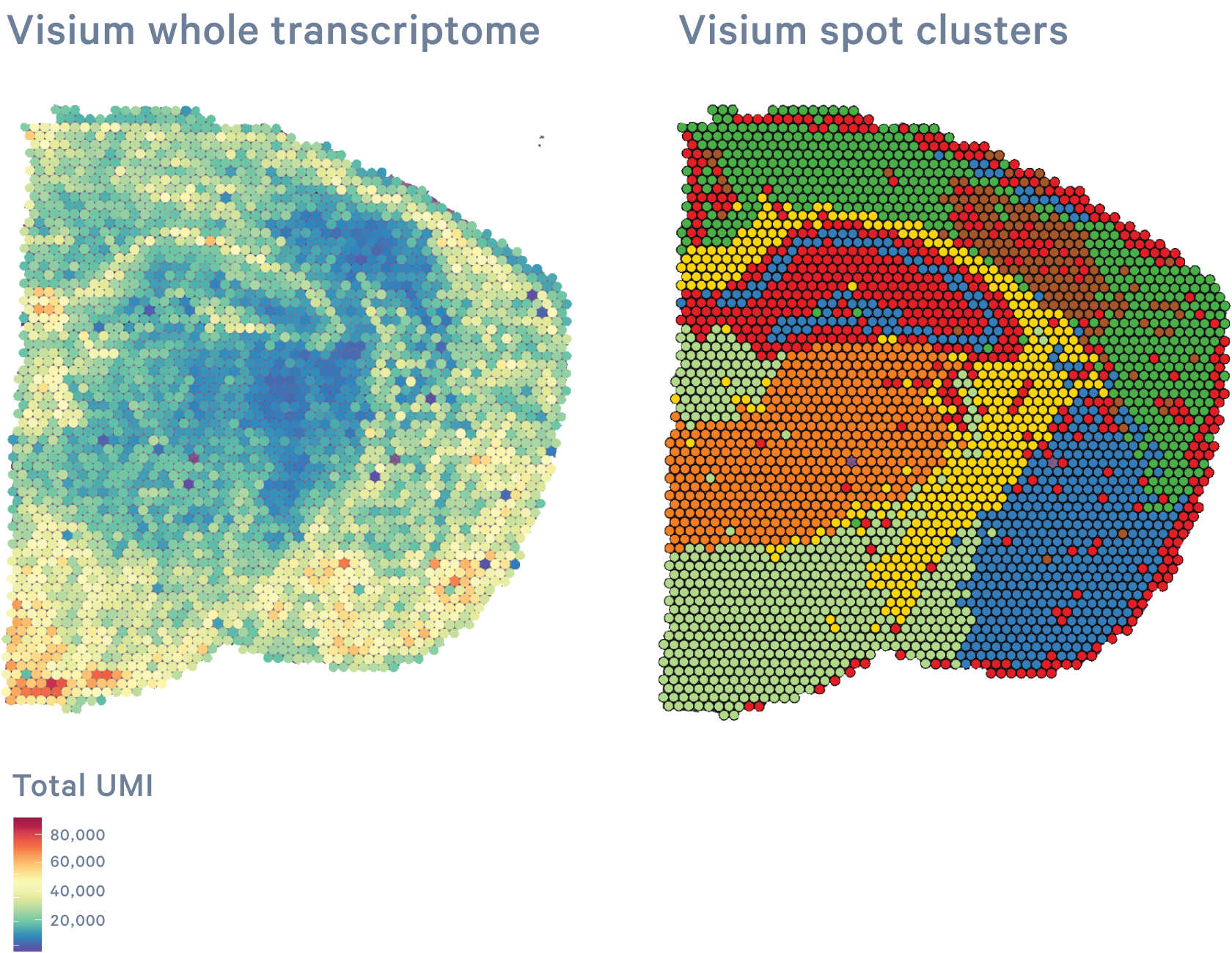
CytAssist Spatial Gene Expression analyzes the whole transcriptome of FFPE, Fresh Frozen, or Fixed Frozen tissue sections in a spatially resolved manner by mapping gene expression over a high resolution microscope image of the tissue.
Documentation
Find resources, from protocols to free-to-use analysis tools for processing human or mouse FFPE, Fresh Frozen, and Fixed Frozen tissues.
Documentation for Fresh Frozen and Fixed Frozen Applications.
Plan ahead by referring to the list of equipment, kits, and reagents needed for the Visium CytAssist Spatial Gene Expression protocols.
Best practices for handling FFPE tissue samples and slides before and after sectioning. Guidance for removing hardset coverslips from archived tissue slides.
Deparaffinize, stain, image, and decrosslink FFPE tissue sections.
For imaging hardware, general image acquisition and analysis guidelines, and example images.
Find a list of probes targeting protein-coding genes in human or mouse transcriptome for the Visium CytAssist Spatial Gene Expression assay.
Construct a sequencing-ready Visium spatial library from your FFPE tissue sections.
Refer to instrument-specific documents for seamless instrument operation.
Sequence your Visium spatial library as per our provided recommendations.
Analyze your Visium spatial data using our pipelines and visualization tools.
Additional Applications
Datasets
Access free, publicly available data from a number of different sample types generated with Visium Spatial solutions
Q&A
Find answers to common technical questions, from sample prep through data analysis, about Visium spatial workflows.
Publications
Read key peer-reviewed research publications offering greater insights into biology, enabled by 10x Genomics products and technologies.