Elucidating the functional domains of a leukemia oncoprotein with single cell CRISPR tiling screens
At our first-ever CRISPR Functional Genomics Symposium, experts in the field shared the latest advances in single cell CRISPR screening. With so many exciting applications to cancer, neurodevelopment, genetic disease research, and more, you don’t want to miss the full series, available on-demand here. And, keep reading to take a closer look at one innovative method presented at the symposium: single cell CRISPR tiling screens, a new approach to perturb coding genes and determine functional domains within the protein product while simultaneously assessing transcriptomic profiles from individual perturbed cells. Learn how a team of cancer researchers from City of Hope used this technique to better understand the functional domains of DOT1L, an enzyme member of a fusion oncoprotein associated with poor prognosis in highly malignant mixed-lineage leukemia.
At our recent CRISPR Functional Genomics Symposium, we heard from leading scientists who are applying CRISPR screening in new and innovative ways to address their biological questions. One active area of technological development in the field is the integration of CRISPR screening with single cell sequencing methods, enabling efficient detection of guide RNA with whole transcriptome profiling of individual edited cells in the same experiment. Benjamin Borgo, PhD, Head of Portfolio Management and Product Development at MilliporeSigma, and a featured speaker at the symposium, noted the potential of this approach:
"...functional genomics has the ability to use ultra high-throughput techniques like DNA sequencing to capture large and highly informative datasets. When we combine these with novel genome engineering tools, it allows for the rapid construction and dissection of gene pathways that govern cellular function and physiology.”
Indeed, innovations on both the CRISPR and single cell sequencing fronts are adding new capabilities to the CRISPR system, including access to multiomic cellular profiles with linked perturbations, providing a full view of the relationship between genotype and phenotype.
Improving research efficiency with single cell CRISPR screens
Innovations in single cell CRISPR screens are not only increasing the depth of insights that researchers can access through their experiments, but also providing greater confidence in the quality of those insights while improving experimental efficiency. Symposium speaker, Dr. David Chen, Assistant Professor, Department of Systems Biology at City of Hope, noted that one disadvantage of traditional CRISPR screens is the lack of certainty about whether a cell is receiving only one guide RNA or multiple, resulting in a mixed bag of perturbations that are challenging to resolve. However, with the ability to track which guide RNAs individual cells are expressing through a sequencing-based readout, cells with multiple guides can be filtered out computationally.
“We always will have some portion of the cells that are receiving more than one guide RNA, and that could increase the concern about the phenotype we observed—is that caused by the combination effect of different guide RNA targeting different genes or different regions in this protein? [...] So to reduce the complexity of the screen, it is an easy solution for us that the cells are expressing only a single guide RNA. And in this case, we also know these guide RNAs are expressed in these single cells.”
Dr. Chen also noted a difference in the timeline of traditional CRISPR screens, which rely on a cell death/cell survival phenotype, compared to the single cell CRISPR screen. While the former requires two to three weeks for cells to grow in culture until significant depletions in the population can be observed, single cell CRISPR screens not only accelerate time to results, but give access to information that a survival readout cannot reveal. For one experiment, Dr. Chen noted:
“...transcriptomic profiling was actually performed three days after the viral transduction. [As] compared to the survival screen, at the same time point, [where] there's no obvious cell number changes, suggesting that transcriptomic profiling may precede the cell end phenotype, including the cell differentiation and the survival change.”
Importantly, this allows scientists to approach the mechanisms they’re studying at an earlier stage after perturbation. Researchers can preserve additional time and labor with the ability to detect CRISPR edits and their resulting transcriptomic effects in the same experiment. By contrast, current CRISPR screening approaches require a separate sequencing step after culturing cells.
We’re just beginning to realize the potential of single cell CRISPR screening, but the ultimate measure of its utility is through the discoveries it is enabling. In the following sections, we take a closer look at single cell CRISPR tilling screens, a new capability within the CRISPR system, and how symposium speaker Dr. David Chen used the screen to study DOT1L, an enzyme involved in driving highly aggressive leukemia.
Exploring the efficacy of a selective DOT1L inhibitor in leukemia
Cancer is, according to its simplest definition, uncontrollable cellular growth (1), leading to the formation of tumors. Ironically, there is a hidden protein machine that precisely controls this deadly dysregulation, resulting in gene expression patterns that drive hyperactive cellular growth or abnormal cellular differentiation. Targeting these co-opted oncoproteins and oncogenes to stop their abnormal behavior thus represents one powerful therapeutic approach to address cancer progression at its source.
Scientists and doctors have taken advantage of this approach in treatment of mixed-lineage leukemia (MLL), a highly aggressive blood cancer that mostly affects children (2). A key driver of this cancer is a fusion protein that activates expression of the A cluster HOX genes, supporting the immortalization of hematopoietic progenitors (3). In a recent clinical trial, scientists targeted one unit of this fusion protein, DOT1L, with a small-molecule inhibitor, then observed the treatment’s effects on cancer progression in a cohort of adult patients with advanced acute leukemias, including MLL (4). Out of 51 enrolled patients, 2 experienced full remission, suggesting DOT1L inhibitors have strong potential in combination with other leukemia treatments.
More study was needed, however, to understand the role DOT1L plays in the fusion complex and thus how to optimally design inhibitors. This led a team of cancer researchers from City of Hope, headed by Dr. Chen, to explore the composition of DOT1L more closely. How does it interact with the fusion complex? What functional domains drive the protein’s activity? How could modulation of the selective inhibition of these domains alter treatment efficacy?
To address these questions, they developed CRISPR tiling screens, a method for high-density CRISPR targeting of coding exons that can identify functional domains in genes (5). With this new approach, Dr. Chen’s group could perturb the gene encoding DOT1L at specific intervals, disrupting the amino acid sequence and so defining the regions of the protein that contribute to its function.
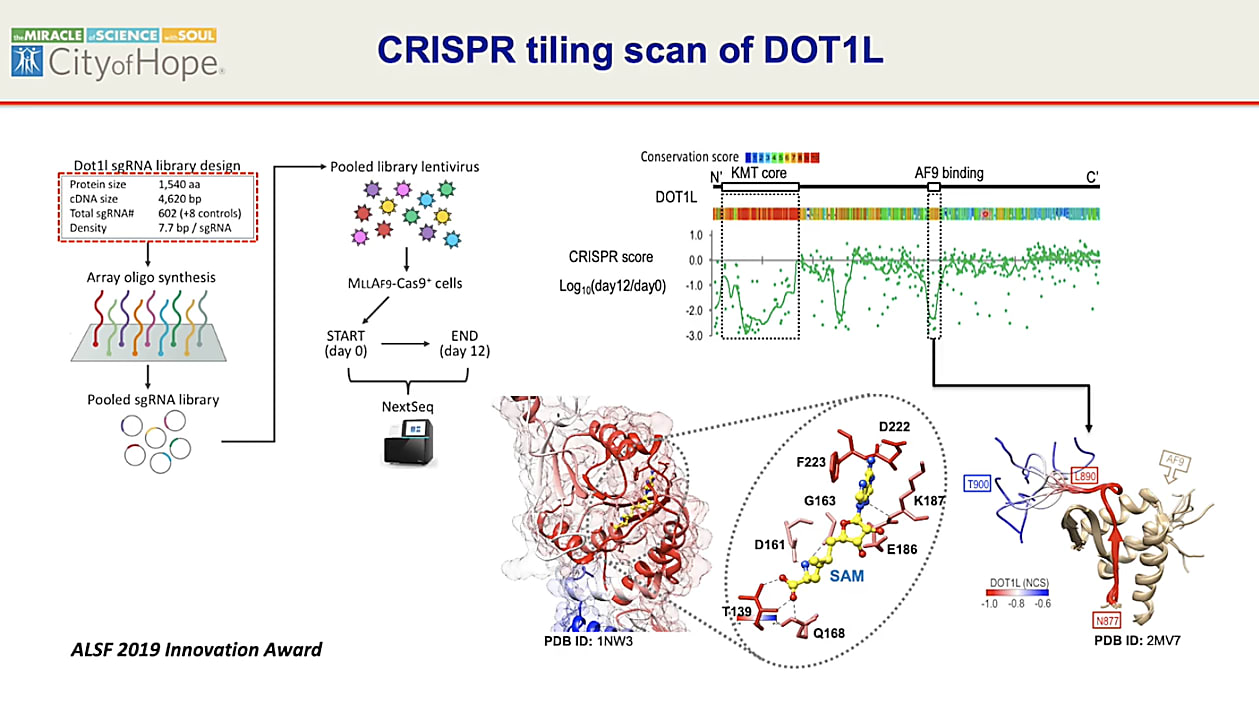
Delivering their guide RNA CRISPR library targeting the DOT1L-encoding gene into MLL cells expressing Cas9, Dr. Chen and his team searched for populations of cells that were more proportionally depleted, suspecting that they were likely carrying guide RNAs that impaired DOT1L function. This survival readout could then point to protein regions that play the most vital role in not only supporting DOT1L function, but also promoting the MLL fusion product.
Although powerful, Dr. Chen noted that there are limitations to this screening method:
“…these types of screens, though powerful, are more limited to the initial mechanistic information than how the cells evolve to a cell death or proliferation phenotype. [...] It's always a desire in the CRISPR genetic screening field that we have an efficient way to link the CRISPR genotype to a phenotype, allowing us to actually see the whole transcriptome.”
Linking leukemic transcriptomic signatures with DOT1L protein functional domains
To advance their experiments with this desired resolution, Dr. Chen’s team then took another approach by turning to single cell CRISPR screens from 10x Genomics. Modifying their CRISPR gene-tiling method to incorporate single cell transcriptomic data, a method they called sc-Tiling, they prepared a 600 guide RNA CRISPR library targeting the entire length of the DOT1L gene in leukemia cells. They observed that seven transcriptionally distinct cell clusters arose from their perturbations, tracking the expression of leukemia-associated genes, including MIXL1, HOXA9, and MEG-01. This analysis revealed a spectrum of cellular differentiation, such that clusters on the right side of their UMAP projection had a more leukemia-associated gene expression profile, while those on the left had a macrophage/myeloid cell profile.
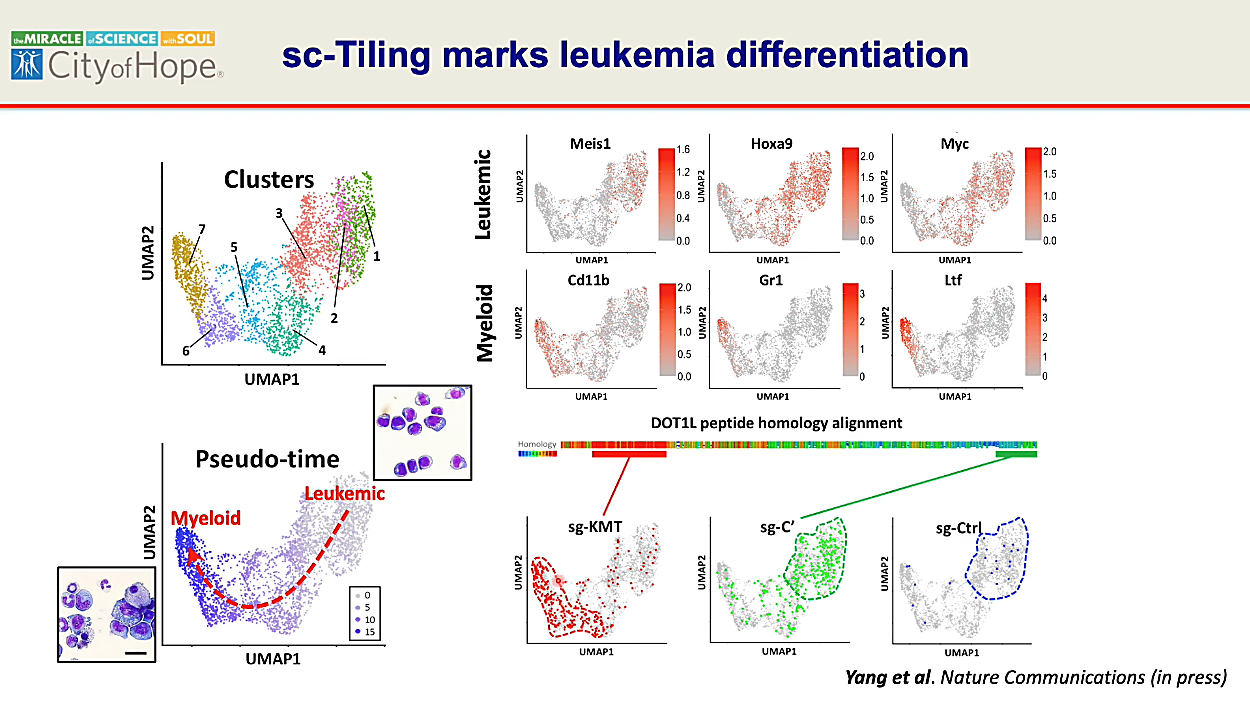
Crucially, with access to guide RNA information and transcriptomic signatures in the same individual cells, the team was able to align perturbed regions of the DOT1L gene with the resulting transcriptomic effects. This revealed that the guide RNAs targeting the DOT1L core domain elicited a myeloid signature, pointing to the importance of that region for DOT1L function within the MLL fusion product. A number of additional functional regions emerged through their analysis of the single cell transcriptomic data across all perturbed cells, including a similar transcriptomic profile between cells perturbed in the core domain and a region near the C-terminal module of the protein, called the R domain.
“The association of the R domain with the core enzyme domain also made us think that the R domain may be involved in other enzymatic activation, and also may modulate the inhibitory therapeutic response to DOT1L inhibitor.”
This insight led the team back to a large patient dataset, to look for clues to the role of this R domain in mediating the therapeutic response to DOT1L inhibitor. They identified a pattern of point mutations in a subregion of the R domain, called R2, which, when cloned into leukemia cells, conferred a significant drug-resistant phenotype requiring higher doses of DOT1L inhibitor to suppress protein activity. Together, their findings suggested a new model of DOT1L function—where R1 and R2 may be required for enzymatic activity, but point mutations in the R2 domain could lead to a hyperactive, drug-resistant state—with implications for therapeutic development of DOT1L inhibitors.
Advancing the frontiers of CRISPR screening
It’s amazing to see the ongoing innovations in CRISPR technology, as scientists use it in new and unexpected ways, uncovering gene function, regulators involved in cell fate determination, mechanisms of disease, and now, protein functional domains. We’d like to thank all of the researchers who contributed to our recent CRISPR Functional Genomics Symposium, including Dr. Chen and his team from City of Hope, for driving this advancement. Even greater possibilities for improved understanding of and therapeutic intervention in major diseases are on the horizon with an integrated view of perturbation and the resulting transcriptomic effects at single cell resolution—and we can’t wait to see them.
Dr. Chen’s presentation, as well as other webinars from the 10x Genomics CRISPR Functional Genomics Symposium, are available on-demand. For more details on single cell CRISPR tiling screens and how this novel technology was used to study DOT1L, you can read the full publication here.
Want to learn more about Chromium Single Cell CRISPR Screening from 10x Genomics? Explore here →
References
- National Cancer Institute. What is cancer? May 5, 2021. https://www.cancer.gov/about-cancer/understanding/what-is-cancer
- Slany RK. The molecular biology of mixed lineage leukemia. Haematologica 94(7): 984–993, 2009.
- Muntean AG and Hess JL. The pathogenesis of mixed lineage leukemia. Annu Rev Pathol 7: 283–301, 2012.
- Stein EM, et al. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood 131(24): 2661–2669, 2018.
- Yang L, et al. High-resolution characterization of gene function using single-cell CRISPR tiling screen. Nat Comm 12: 4063, 2021.
