Stories of progress, potential, and possibilities: Recapping AAIC 2022
Alzheimer’s disease is a thief of many things: joy. Time. Memory. It is prolific, with estimates that more than 30 million people worldwide suffer from this illness (1). Most of all, it is cruel, stealing not just precious memories of the past but preventing the formation of new ones.
Those who suffer from Alzheimer’s disease may forget, but we—their friends, family, and loved ones—remember. And a select few of us are privileged to make a career out of working towards understanding, and developing an eventual treatment for, this terrible disease.
Alzheimer’s disease, however, is not an easy disease to study. More than a century has passed since its characterization, but disease-modifying treatments remain elusive. However, it’s been less than ten years since single cell RNA-sequencing (scRNA-seq) and spatial transcriptomics have started making waves, and the discoveries fueled by these technologies are extraordinary.
At AAIC 2022, more than 5,000 members of the Alzheimer’s disease (AD) field came together to share their latest insights, findings, and the tools they used to generate them. Read the recap of some of the key stories below, including the role of microglia in AD in immunometabolism, the characterization of the APPSWE mouse model with spatial transcriptomics, and upcoming technological advancements that will allow researchers to push the field further than ever before.
Award-winning research: Honoring work on neuroinflammation, metabolic dysfunction, and APOE
To honor novel and innovative findings in single cell and spatial transcriptomics, 10x Genomics features a poster competition each year at AAIC. This year, the winner was Nicholas Devanney, a graduate student at the University of Kentucky.
Nicholas’ poster centered on the microglial function of apolipoprotein E (APOE)—the strongest genetic risk factor for Alzheimer’s disease (AD)—and its role in AD-linked neuroinflammation and metabolic dysfunction. While APOE and the transcription factor Hif1a are thought to drive the shift from “normal” microglia to an AD-specific damage-associated microglia (DAM) subtype, prior studies have focused only on mouse APOE.
Nicholas addressed this shortcoming with mice where the endogenous APOE was replaced with the human version of APOE3 or APOE4 (APOE-TR mice). scRNA-seq of glia-enriched samples using Chromium Single Cell Gene Expression of APOE4 mice revealed metabolic pathways, carbon metabolism, and glycolysis/gluconeogenesis as most impacted in glia. In microglia specifically, genes upregulated in aged and APOE4 mice were enriched for DAM and MgND profiles; however, APOE was confined to a single cluster characterized by significantly higher Hif1a activity and glycolysis pathway scores compared to all other clusters. They also noted that the cellular metabolism of APOE4 microglia seemed to reflect significant alterations in glycolysis and mitochondrial respiration compared to APOE3 microglia, consistent with neuroinflammation and metabolic dysfunction.
But these were in “normal” mice; what happens in a model of AD? Using spatial transcriptomics in brains from APOE-TR mice crossed with the 5xFAD strain (a mouse model of AD), they identified an APOE4-specific cluster strongly enriched for metabolic pathways. This cluster was present predominantly in hippocampus and cortex, with multiple DAM and neurodegenerative microglia-associated genes enriched in both of these brain regions.
Taken together, Nicholas’ work proposes a model in which APOE4-induced metabolic deficits contribute to one of the earliest hallmarks of AD—neuroinflammation. In addition, these findings provide not just a mechanism tying these two phenomena together, but also link it to a major genetic risk factor for AD, providing a new avenue of inquiry for therapeutic development.
We’d like to take this opportunity to congratulate not only Nicholas for his work, but also the two runners-up in our poster contest:
- “Distinct cerebrospinal fluid immune perturbations in healthy brain aging and cognitive impairment” (Natalie Piehl, Northwestern University) Read now →
- “Impact of PLCG2 AD variants on microglial biology and disease pathogenesis” (Andy Tsai, Indiana University School of Medicine) Read now →

Putting the “where” in the “what:” Spatial transcriptomics in a mouse model of AD
While the anatomy and progression of plaques and tangles in AD is well-characterized, the underlying mechanisms of these phenomena remain elusive. This is partly because, while most technologies examine the “what” of gene expression changes and their cellular basis, they are unable to capture the “where” of their anatomical basis. Our poster, “Spatially resolved transcriptomics in the APPSWE [Tg2576] mouse model of Alzheimer’s disease,” demonstrated how researchers can address this shortcoming with spatial transcriptomics.
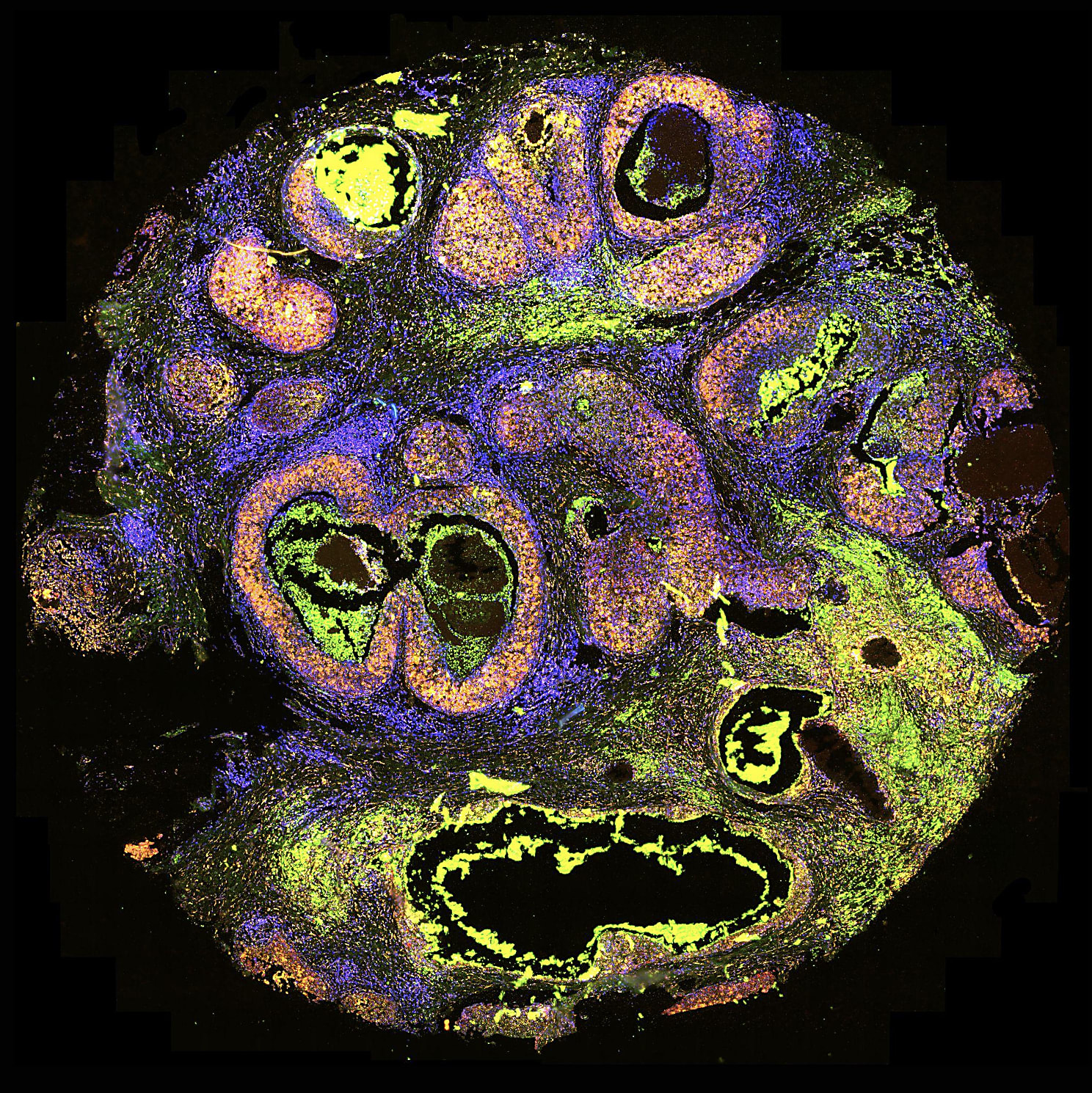
Using the APPSWE mouse model of AD, our team took serial sections of fresh-frozen brain tissue and analyzed them with Visium Spatial Gene Expression. This whole-brain spatial transcriptomics approach not only recapitulated mouse brain anatomy (Figure 1) but mapped key areas of relevance to AD, including hippocampus and cortex.
When looking at gross gene expression changes across the entire brain, three hormone-coding transcripts—Pmch, Hcrt, and Trh—were significantly differentially expressed in APPSWE mice versus controls. The most dramatic changes occurred in a specific brain region (the hypothalamus), while other areas of the brain had comparatively low expression, underscoring the importance of considering neuroanatomical regions in identifying new prospective biomarkers and genes of interest. Finally, our team highlighted the ability to merge immunofluorescence-based protein detection with spatial transcriptomics on the Visium slide, showing a link between increased Pmch expression at the transcript level with Mch at the protein level.
Combining approaches and showcasing what’s to come
Sometimes it helps to examine the same problem from multiple angles. Hongjun (Harry) Fu, PhD (Ohio State University) demonstrated this in his presentation, where he noted that, while scRNA-seq is valuable, “This procedure unavoidably causes loss of positional or spatial information of the cells.” His talk recapped his previous work showing that excitatory neurons in AD show a tau homeostasis signature and exhibit both cellular and regional vulnerability. He then discussed his latest work, where he combined snRNA-seq with Visium Spatial Gene Expression to explore the underlying mechanisms of selective cellular vulnerability in AD. This multifaceted approach not only identified multiple genes dysregulated in AD, but found genes that were disrupted in both a layer- and pathology-specific manner. The data he presented is from a manuscript currently under review, so keep an eye out for his publication (and a more in-depth discussion of his work!) in the near future.
After Dr. Fu’s talk, our product marketing manager, Jyoti Sheldon, showcased two of our technologies that will help researchers push studies like these even further: the Visium CytAssist instrument, and the Xenium In Situ platform (both of which were on display in our booth!). On the spatial transcriptomics front was CytAssist, which further simplifies the Visium workflow and enables the use of presectioned FFPE tissues on the Visium platform. The features she highlighted are ideal for researchers with access to large stocks of archival FFPE tissues from patients with AD or related dementias that seek to better understand transcriptomic dysregulation with pathological context, identify biomarkers, and more.
The other new technology she highlighted was our upcoming Xenium In Situ platform, which will allow customers to assay hundreds of gene targets at subcellular resolution at both the RNA and protein level (Figure 1). This technology will be compatible with both fresh-frozen and FFPE tissues for identifying key biological events, and is well-positioned for high-throughput studies of larger cohorts (such as those used in drug screening and interaction studies). We’re gearing up for more on this, so stay tuned!
Capping off and moving forward
Conferences like AAIC 2022 are both humbling and inspiring: humbling for showing how far we have to go; inspiring for showing how far we’ve come—and where we’re going next. Findings from researchers like Dr. Fu and Nicholas Devanney, novel advances, and the insights being gleaned from single cell and spatial technologies are paving the way forward in the field, and moving towards disease-modifying therapeutics for AD faster than ever before. We hope to see you all again next year, and can’t wait to see what you do in the upcoming months!
In the meantime, check out our Alzheimer’s research resources and discover what others have done with single cell and spatial transcriptomics tools in our research highlights, explore potential applications in the application note and infographic, and more.
References:
- Dementia. Who.int. https://www.who.int/news-room/fact-sheets/detail/dementia
