The Tadpole, the Mouse, and the Worm: Studying Development in Model Organisms
Through the study of model organisms like yeast, worms, flies, and zebrafish, scientists have made fundamental discoveries about developmental biology. They’ve learned how fertilized eggs develop into complex, segmented organisms and what genes control this process. They’ve even been able to track the cellular lineages of entire organisms. Nobel Prize winner, John Sulston used light microscopy to learn that C. elegans, a nematode and favored model organism, is composed of exactly 959 somatic cells. This discovery enabled researchers to begin addressing questions about how development happened at the level of single cells.
Today, researchers continue to build on these fundamental discoveries, adding single cell transcriptional profiling to the mix of techniques traditionally employed to study development. Here we review some of the discoveries that are shaping our understanding of the cellular, molecular and temporal mechanisms underlying regeneration, cell type specification and organ formation in model organisms — and that are giving us a more complete picture of development.
Novel cell type coordinates regeneration of tadpole tails
African Clawed Frogs, or** Xenopus laevis, are known for their remarkable regeneration abilities as tadpoles. In the ongoing effort to understand how regeneration works, including what cells and genes coordinate this activity, researchers from the University of Cambridge performed single cell RNA sequencing on X. laevis **tails at various stages after amputation in both regeneration-competent and -incompetent tadpoles. Evaluating single cell data from over 13,000 cells, they found a previously unidentified cell type of the epidermis that was observed only after tail amputation in regeneration-competent tadpoles. This cell type expressed major growth factors and instructive signals that support regeneration, including Wnt5a; and subsequent GFP staining and immunolabeling confirmed these novel, regeneration-organizing cells (ROCs) relocalize to the amputation plane, forming the wound epidermis. Further study strengthened the cell’s identity: for example, transplantation of ROC-containing grafts induced ectopic outgrowths in early embryos. Taken together, these findings suggest that ROCs are crucial to the regenerative response, and may reflect a conserved feature of appendage regeneration in other species (1).
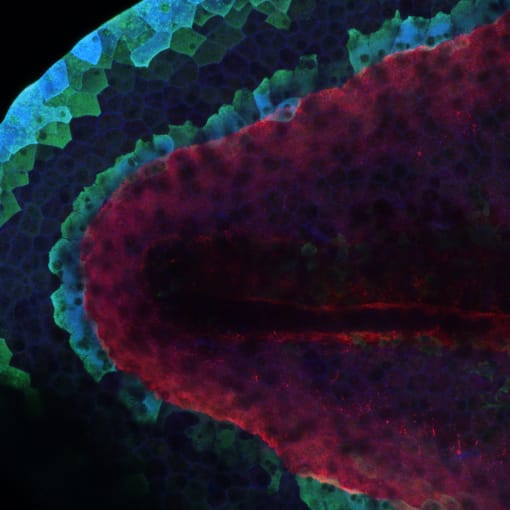
Tracking endodermal differentiation in the mouse embryo
A major goal of developmental biology research includes characterizing the order and nature of cellular events that give rise to complex organs from preliminary germ layers. Recently, researchers from the Sloan Kettering Institute sought to understand what transcriptional changes differentiate two endoderm cell populations in mouse embryos—extra-embryonic endoderm (that will form the placenta) and embryonic endoderm (that will form internal organs like the gut). They performed single cell RNA sequencing on 112,217 cells, isolated sequentially from mouse embryos spanning the blastocyst through midgestation developmental stages. Transcriptional profiling allowed them to model the differentiation trajectories of these cells, elucidate when and how fate decisions occur, and determine the embryonic or extra-embryonic origins of cells that compose the emerging gut tube. This comprehensive survey of endoderm differentiation within the mouse embryo may also support future studies that seek to understand the developmental origins of cancers that arise in internal organs (2).
Read more about this discovery →
A molecular atlas of C. elegans embryogenesis
As John Sulston discovered, C. elegans are conducive to whole-organism studies because they are composed of a limited number of cells consistently produced in the same sequence of cell divisions. Scientists from the University of Washington and the University of Pennsylvania have revisited the single cell map of C. elegans. Using scRNA-seq and imaging techniques with fluorescent reporter genes, they reconstructed the temporal sequence of gene expression changes that accompany nearly every cell division in the worm’s embryonic cell lineage. This has allowed researchers to study the role of transcriptional changes in driving cell-fate specification during development, and may reveal specific genes and regulators that serve as gates to subsequent levels of terminal differentiation (3).

The fundamental resolution offered by 10x Genomics technology complements established techniques for studying development. These papers demonstrate how using these tools, together with the experimental advantages of model organisms, can provide access to unexpected details about cell growth and differentiation.
And they demonstrate that model organisms are just plain cool.
You can learn more about the single cell RNA sequencing methods that aided these discoveries here →
- Aztekin et al., Identification of a regeneration-organizing cell in the Xenopus tail. Science. 364, 653–658 (2019)
- Nowotschin et al., The emergent landscape of the mouse gut endoderm at single-cell resolution. Nature. 569, 7756 (2019).
- Packer et al., A lineage-resolved molecular atlas of C. elegans embryogenesis at single-cell resolution. Science. 365, 1971 (2019).
