Uncovering the Role of Hand2 in Congenital Heart Defects
**Guest author: jamie-schwendinger, Scientific Project Manager at 10x Genomics **
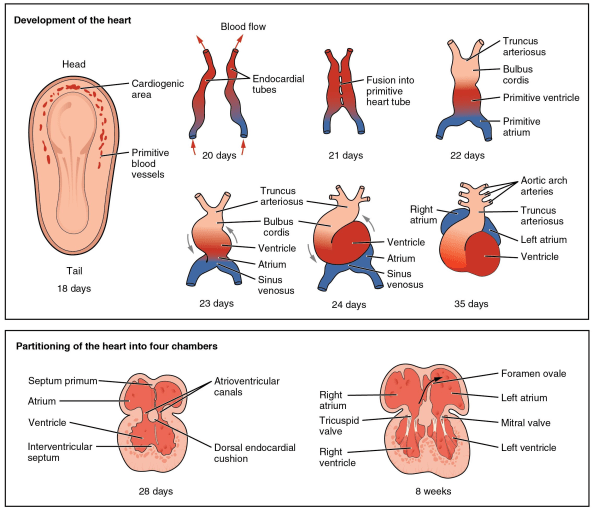
During heart formation, the cardiac mesoderm migrates to the heart field where it forms the cardiac crescent. One day later (in a mouse), these cells have formed a tube, through which blood will flow. A day after that the tube loops, in order to begin forming the characteristic 4-chamber structure of the heart and the outflow tract, a transient structure that gives rise to the aorta and pulmonary trunk.
If this process sounds complicated, that’s because it is. Indeed, congenital heart defects are the most common cause of birth defects, affecting nearly 1% of live births in the United States (cdc.gov).
To understand how and why congenital heart defects happen, and how best to intervene, PhD Candidate Yvanka de Soysa and researchers at the Gladstone Institute of Cardiovascular Diseases turned to single cell RNA sequencing to characterize heart development in mice (1). Rather than measuring gene expression changes averaged across an entire tissue, single cell sequencing enables scientists to measure RNA transcription cell by cell, resolving the local differences that often dictate how developmental processes unfold. However, going from even this astonishingly detailed transcriptomic map to a molecular mechanism still requires considerable resourcefulness.
To get at these molecular mechanisms, de Soysa et al. first microdissected the heart region of mouse embryos at three time points, spanning the duration of heart tube formation and looping. Using the Chromium Single Cell Gene Expression Solution, they sequenced over 36,000 individual cells, identifying 21,366 mesodermal and neural crest cells that contributed to heart formation. These cells were grouped into different cell populations and placed along a developmental trajectory using pseudotemporal ordering, a method that tracks the progression of single cells through an inferred process (like development or differentiation).
Armed with this cellular coordinate system, collaborators in the Computational Biology Group at the Luxembourg Centre for Systems Biology used a computational prediction method to identify transcription factors likely responsible for specification of distinct cardiac structures (2).
Surprisingly, this method predicted that the transcription factor Hand2 specifies the outflow tract, but not the right ventricle. However, deletion of Hand2 results in a well-known phenotype where the right ventricle is drastically reduced in size. How, then, could Hand2 not be responsible for specification of the right ventricle?
To investigate, de Soysa and colleagues compared their wild type single cell gene expression data to that of Hand2 null mutants. They found that, as predicted, Hand2 mutants neither specified nor formed cells of the outflow tract, but did specify right ventricle cells. Pseudotemporal ordering, however, showed that the right ventricle cells in the mutants never fully differentiated. Further, Hand2 mutants at the latest measured time point of embryogenesis had equivalent numbers of right ventricle-like cells, but, because those cells failed to properly differentiate, they never migrated to the correct location and the right ventricle could not properly form. This finding suggests that the transcription factor Hand2 is not necessary to specify right ventricle cells, but is necessary to bring these cells through terminal differentiation.
With the power of single cell sequencing, scientists were able to define a molecular mechanism that disrupted formation of multiple heart structures, and demonstrate the broad and unexpected impact of a single mutation in a tiny population of cells. Moreover, their use of single cell data enabled three key advances that would be impossible using bulk RNA sequencing. Scientists were able to:
- Identify distinct cell populations and their full transcriptome in different regions of the heart
- Elucidate multiple progenitor/progeny relationships using pseudotemporal ordering
- And characterize key transcription factors that gate binary decisions during development
This study reflects the exciting possibilities of single cell sequencing technology to advance our understanding of development in both healthy and disease states, across biological systems and organ models. By understanding the cellular and molecular mechanisms that cause congenital heart defects, scientists hope to be able to prescribe preventative strategies for people genetically at risk for this disease. As Dr. Deepak Srivastava, the lead author of the study, explains in a statement, "The ultimate goal is to create [...] public health measures that could reduce the overall incidence of birth defects through prevention. But first, we have to know where and how to intervene."
- De Soysa, Ranade, Okawa, et al. Single-cell analysis of cardiogenesis reveals basis for organ-level developmental defects. Nature. 572, 120–124 (2019).
- Okawa, del Sol. A computational strategy for predicting lineage specifiers in stem cell subpopulations. Stem Cell Res. 15, 427-34 (2015).
