Untangling spider silk secrets with multidimensional analysis
We hate to break it to all the Spider Man fans out there, but humans aren’t able to spin these webs just yet—whether it’s for medical, commercial, or superhero purposes. Nature still does it best.
However, with the aid of single cell and spatial transcriptomics technology, scientists are defining the molecular biological mechanisms of silk production in one amazing species, the gold-silk producing Trichonephila clavata, also fittingly known as the golden orb-weaving spider.
Detailed insights into the cellular composition and structural organization of the organ behind dragline silk production in orb-weaving spiders, the major ampullate (Ma) gland, build on existing knowledge of the physical and material properties of super strong spider silk and hold promise to inform human efforts to generate optimized synthetic silk for our own needs—like swinging from rooftops (1).
The spider Ma gland, not Spider Man, is the superhero of silk production
In their recent Nature Communications paper (1), a team of scientists from Southwest University, Chongqing, China, described a multiomic study of the golden orb-weaver’s major ampullate (Ma) gland. This gland helps the spider produce dragline silk, which is used for its notorious orbed webs, and even for “ballooning,” a means of air travel for the spiders (1, 2).
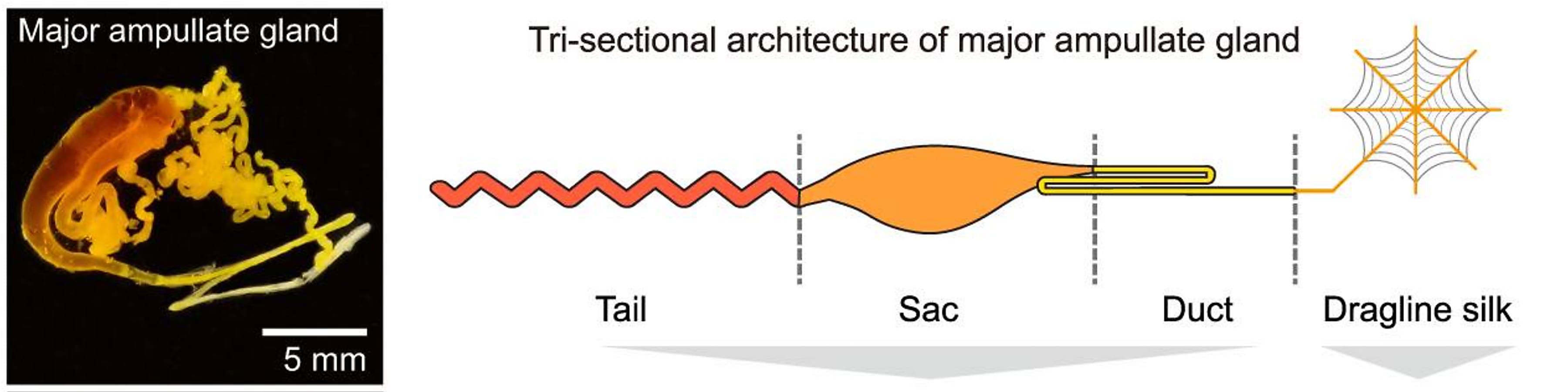
The Ma gland is composed of three distinct segments: the tail, sac, and duct. Using bulk RNA-seq, the team characterized the transcriptional features of each segment, hoping to trace the origins of the 28 protein components of dragline silk and define a molecular mechanism of silk production. They found, based on transcriptomic and proteomic analyses, the tail and sac have similar molecular functions as the major silk-secreting segments. Specifically, the tail was enriched for genes involved in the synthesis of organic acids, while the sac was enriched for genes related to the synthesis of lipids. Genes in the duct were predominantly associated with ion (Ca2+ and H+) exchange and chitin—the main component of a spider’s exoskeleton—synthesis (1).
A mechanism of silk production comes into focus with cellular and spatial insights
To establish a cellular mechanism of silk production, the team used Chromium Single Cell Gene Expression technology to identify the cell types that compose the Ma gland. They analyzed 9,349 single cells, and identified ten unique cell clusters with various molecular roles based on gene expression. Some of these populations included Ma gland origin cells, and multiple cell types responsible for production of the major protein components of the silk, which were sourced to the tail and sac segments. They also identified lipid synthesis cells, chitin synthesis cells, and pH adjustment cells, among others (1).
The team ordered the developmental trajectories of these cell populations, revealing a continuum of cells starting from Ma gland origin cells which clustered in the tail and branched off into the remaining two major segments, the sac and duct. Additionally, using Visium Spatial Gene Expression analysis, they mapped the spatial organization of cell populations within the Ma gland, focusing on the silk protein-secreting cells. This confirmed their localization within the tail and sac segments.
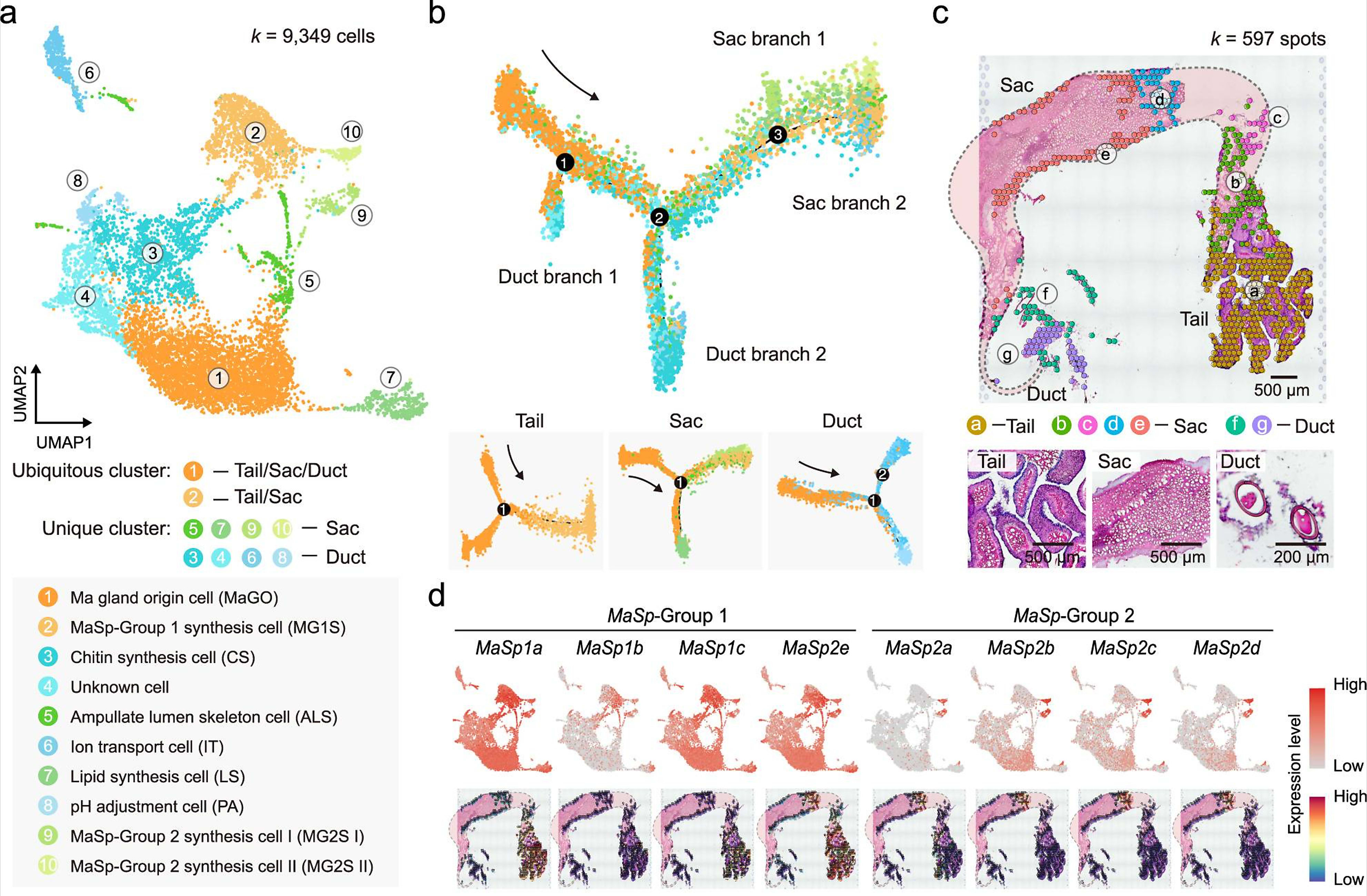
Taking their single cell and spatial datasets together, the research team was able to further refine the molecular function of unique clusters, classifying single cell and spatial transcriptomics clusters into ‘sets’ based on alignment to 35 segment-specific gene expression signatures. This functional map established specific roles for single cell clusters—such as cluster 2, which was shown to play a role in organic acid metabolic process and oxidoreductase activity—and simultaneously established their location within specific regions of the gland tissue (1).
These detailed insights not only provide a mechanistic explanation for the development of the Ma gland, but also resolve a long-held scientific debate over the cellular composition of the gland, including the number of unique cell types and which cells are responsible for making the proteins that contribute to the amazing strength and elasticity of dragline spider silk.
Unraveling the secrets of complex animal—and human—biology
Nature continues to surprise and outdo us. As scientists use high-resolution tools to delve deeper into the complex biological mechanisms that enable the stunning features of the natural world, like the orb-weaver’s web, who knows what we’ll discover next. Whether it’s the underlying biology of the octopus’ camera-style eyes, neural regeneration in axolotls, or embryonic development in rabbits, an emerging model organism, there’s still so much to learn.
But it’s not just the natural world that has untold secrets. Human biology, and how it can go awry in development and disease, must come into greater focus as well. Single cell and spatial tools, and their combined analytical power, have huge potential to support a new wave of discovery into how we work, with implications for translational medicine and clinical improvements.
Keep exploring the human discoveries that single cell and spatial tools have supported with our recent blog series, When single cell meets spatial:
- Researchers uncover how people with paralysis may walk again
- Novel lung immune niche provides long-term antibody hideouts
- Spatial and single cell tools open the black box of early human development
References:
- Hu W, et al. A molecular atlas reveals the tri-sectional spinning mechanism of spider dragline silk. Nat Commun. 14: 837 (2023). doi: 10.1038/s41467-023-36545-6
- Trichonephila clavata. Updated: March 23, 2023. https://a-z-animals.com/animals/joro-spider/
Social share image credit Micha L. Rieser.
