For libraries made with Single Cell Gene Expression v4 or Immune Profiling v3 chemistries, check the
Supported Librariestable to confirm compatibility with the latest Cell Ranger version.
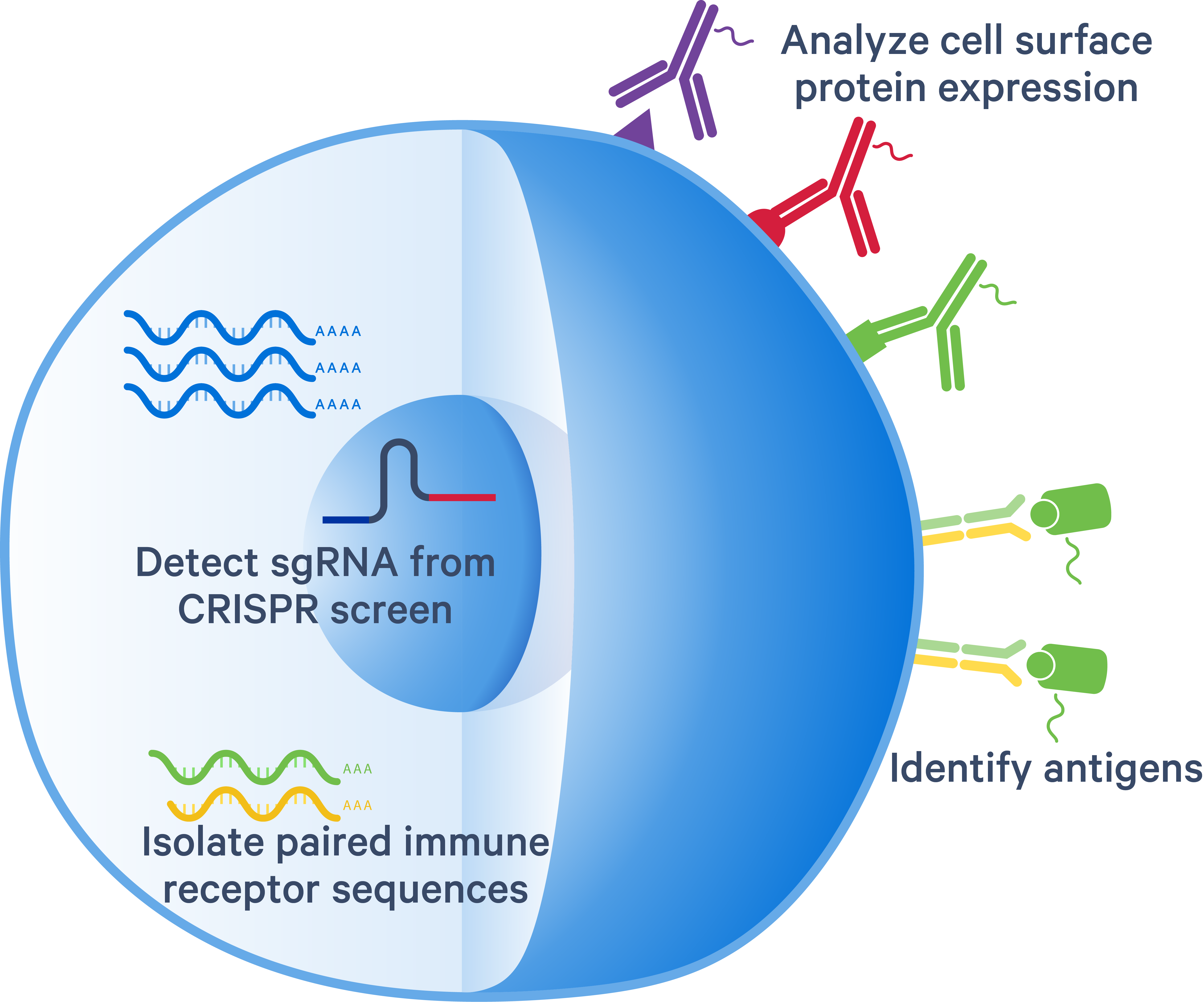
Feature Barcode technology is a method for adding extra layers of information to cells by running single cell gene expression in parallel with other assays.
-
Antibody Capture (cell surface protein measurement): A method to label cell surface proteins using a specific protein binding molecule, such as an antibody conjugated to a Feature Barcode oligonucleotide (e.g. BioLegend TotalSeq™–B Antibodies).
-
CRISPR Guide Capture: A method to analyze gene expression changes caused by the presence of CRISPR perturbations. The assay captures the full transcriptome and transfected guide RNAs from the same cell to correlate changes in the transcriptome to CRISPR perturbations.
-
3' Cell Multiplexing: A method to label a given cell (or nuclei) sample with a molecular tag and subsequently pooling this sample with other labeled samples. Please see the What is Cell Multiplexing? page for assay and data analysis information.
-
5' Antigen Capture: Chromium Single Cell 5’ Barcode Enabled Antigen Mapping (BEAM) is a method to rapidly map paired T and B cell receptor sequences to their corresponding antigens. BEAM is unsupported with the GEM-X chemistry; therefore, Cell Ranger does not support BEAM/Antigen Capture libraries produced with this chemistry..
-
For background on Feature Barcode assay schemes and terminology, see the Feature Barcode Glossary.
-
To learn how to use Cell Ranger with Feature Barcode data, see the Feature Barcode Analysis page for Antibody Capture or CRISPR Guide Capture.
-
To learn how to describe your Feature Barcode reagents as inputs to the
cellranger countpipeline, visit Feature Barcode Reference page. -
For an additional tutorial on how to perform analysis with multiple library types (Single Cell V(D)J + 5′ Gene Expression + Feature Barcode technology), visit the Integrated Analysis section.
-
To learn how to describe Feature Barcode reagents as inputs to the
cellranger countpipeline, visit the Feature Barcode Reference page. -
To learn how to view Feature Barcode data in Loupe Browser, see the Feature Barcode Tutorial.
-
For assay-related resources (User Guides, Demonstrated Protocols, Technical Notes, SDS), see additional applications pages for Single Cell Gene Expression, Single Cell Immune Profiling, or Flex.