Space Ranger v3.0 and later can analyze Visium HD Spatial Gene Expression datasets, and v4.0 introduced support for Visium HD 3' Spatial Gene Expression. The spaceranger count pipeline supports the analysis of both.
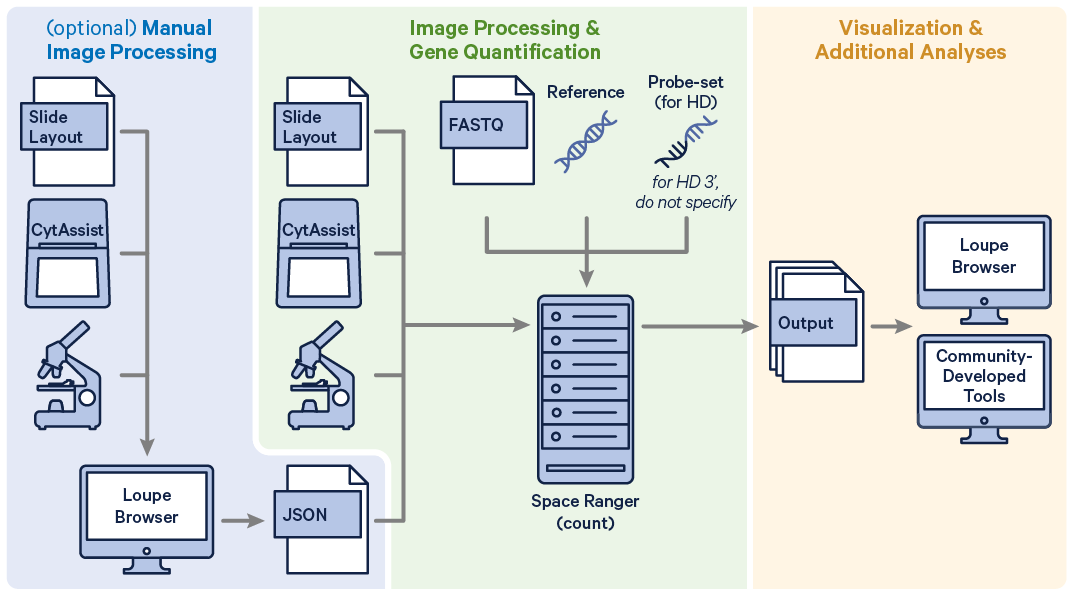
Before running the pipeline, check that you have the following inputs prepared:
- The corresponding CytAssist image in
TIFFformat (--cytaimage). - Microscope image (optional) in either
TIFF,QPTIFF,BTF, orJPEGformat:--imagefor a brightfield microscope image.--darkimagefor a dark background fluorescence microscope image.--colorizedimagefor a composite colored fluorescence microscope image.
- Slide parameters specified by:
--slide&--areaifspacerangerhas access to internet (optional with CytAssist metadata).--slidefile,--slide&--areaifspacerangerhas no access to the internet (the slide layout file must be directly downloaded.--unknown-slideif Visium slide details are unknown.
- The reference transcriptome (
--transcriptome). Human and mouse references are available for download, or build a custom reference. - The probe set CSV (
--probe-set). These files can be found in theprobe_setsdirectory in the Space Ranger software package or downloaded here. When analyzing Visium HD 3' data, do not specify this parameter. - In most cases, Space Ranger will perform automatic image to fiducial alignment and tissue detection. When automatic alignment fails, you must also run manual alignment in Loupe Browser and specify the alignment JSON file with the
--loupe-alignmentoption.
The following command assumes all inputs are in the current working directory; change paths as needed. Run a separate instance of the spaceranger count pipeline on each capture area.
spaceranger count --id=hd_count \
--transcriptome=/path/to/refdata-gex-GRCh38-2020-A \
--fastqs=/path/to/fastq \
--probe-set=/path/to/Visium_Human_Transcriptome_Probe_Set_v2.0_GRCh38-2020-A.csv \
--slide=H1-YD7CDZK \
--area=A1 \
--cytaimage=/path/to/CAVG10539_2023-11-16_14-56-24_APPS115_H1-YD7CDZK_A1_S11088.tif \
--image=/path/to/APPS115_11088_rescan_01.btf \
--create-bam=false
--probe-set.--create-bam argument to be set to true or false. Here, we set it to false to save disk space and computation time. BAM files can be useful for long term data archiving, troubleshooting (especially mapping), and other reasons.--slide and --area options if these metadata can be obtained from the CytAssist image. For a complete list of arguments and options, see the Command Line Argument Reference, or run spaceranger count --help.
Space Ranger v4.0 and later includes support for nucleus and cell segmentation, and downstream secondary analysis based on the segmented results, in the spaceranger count pipeline. By default, the pipeline will perform cell segmentation automatically as long as an H&E tissue image is provided using the --image option (fluorescence images are not supported). Users now have the following options:
- Users can override Space Ranger's default nucleus segmentation algorithm by providing their own nucleus segmentation mask with the
--custom-segmentation-fileoption. If this is provided tospaceranger count, the--nucleus-expansion-distance-micronoption must also be set. Custom segmentation masks can be generated using third party tools or thespaceranger segmentpipeline (also new in v4.0). - For exceptionally large cells (expected diameter >40 µm), users can specify the maximum nucleus diameter with the
--max-nucleus-diameter-pxoption. - Space Ranger's segmentation algorithm can be disabled altogether by setting
--nucleus-segmentation=FALSE.
For more information, see the spaceranger segment, Space Ranger Commands and the Image Processing Algorithms pages.
A successful spaceranger count run concludes with a message similar to this:
Outputs:
- Run summary HTML: /outs/web_summary.html
- Outputs of spatial pipeline:
Slide image annotated with fiducial alignment: /outs/spatial/aligned_fiducials.jpg
Slide image annotated with detected tissue: /outs/spatial/detected_tissue_image.jpg
...
2024-02-26 18:00:10 [perform] Serializing pipestance performance data.
Waiting 6 seconds for UI to do final refresh.
Pipestance completed successfully!
To continue, see Understanding Space Ranger Outputs. It is recommended to check the web_summary.html file first to assess quality control metrics.