The resegment pipeline allows you to generate a new segmentation of the DAPI image space by rerunning the Xenium Onboard Analysis (XOA) segmentation algorithm with modified parameters.
Adjustable parameters:
- Nucleus expansion distance (e.g., to make a nucleus-only count matrix (set to distance to 0 µm) or to use a more conservative expansion distance).
- Minimum DAPI intensity (e.g., to rescue dim nuclei that are filtered by default).
The input files for resegment are:
- The Xenium output bundle generated by the Xenium Onboard Analysis pipeline (see Input overview).
The xeniumranger resegment command line arguments are described on the Xenium Ranger command line arguments page.
Here are some example uses of the resegment pipeline. Make sure to edit the paths to input/output files in the examples below.
If you have QC'd the nucleus segmentation in Xenium Explorer and want to adjust the nuclei expansion distance (default: 15 µm) without altering nuclei detection, use the following arguments:
xeniumranger resegment --id=demo \
--xenium-bundle=/path/to/xenium/files \
--expansion-distance=5 \
--resegment-nuclei=false
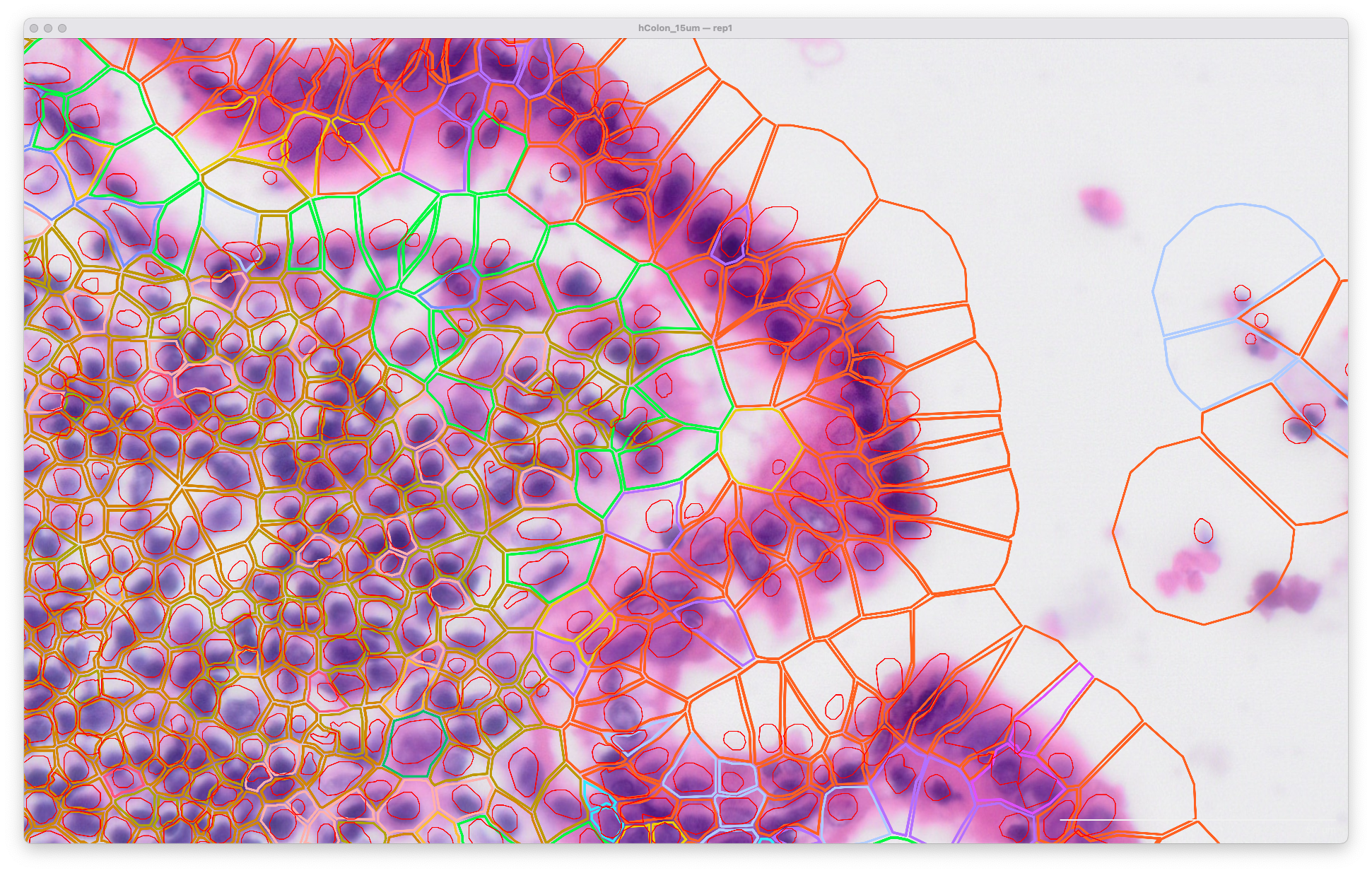
Viewing the original output in Xenium Explorer, the cell and nuclei outlines look like this at the default 15 µm expansion distance for segmentation of cells in the lumen of the human colon:
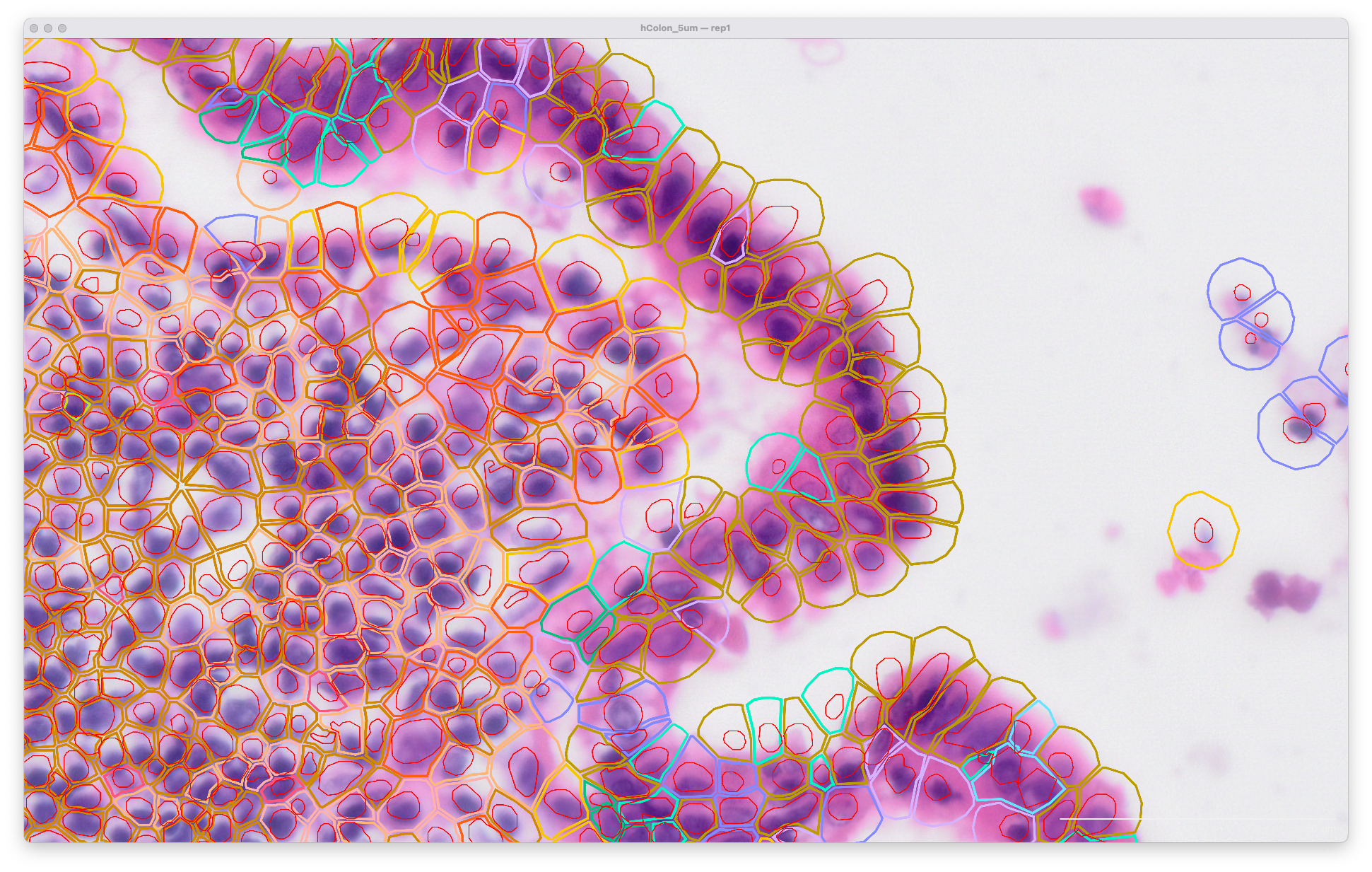
The same area of the resegment analysis output with 5 µm expansion distance looks like this:
The XOA version updates may include improvements to the nucleus segmentation model (i.e., in v1.3, see release notes). If you want to adjust nuclei expansion and use the XOA segmentation model that corresponds to the Xenium Ranger version you are running (i.e., v1.6), use --resegment-nuclei=true (default). This scenario requires the latest version of Xenium Ranger (i.e., if the input data is from XOA v1.1, run with Xenium Ranger v1.6 to get segmentation results generated by the XOA v1.6 pipeline).
xeniumranger resegment --id=demo \
--xenium-bundle=/path/to/xenium/files \
--expansion-distance=10 \
--resegment-nuclei=true
The DAPI filter allows you to increase or decrease the amount of nuclei data used for reanalysis by adjusting a pixel intensity threshold. Pixel intensity is measured in units of photoelectrons, as seen in the morphology images.
For datasets where the DAPI signal is clearly dimmer across the entire imaged area, you can adjust the minimum peak intensity to use more nuclei in the DAPI image for nucleus segmentation (default: 100 photoelectrons). This parameter is not designed for fine-tuning datasets that have only a few missing nuclei. In this example, any nucleus for which 95% or more of its pixel intensity is less than an intensity threshold of 50 photoelectrons will be removed.
xeniumranger resegment --id=demo \
--xenium-bundle=/path/to/xenium/files \
--dapi-filter=50
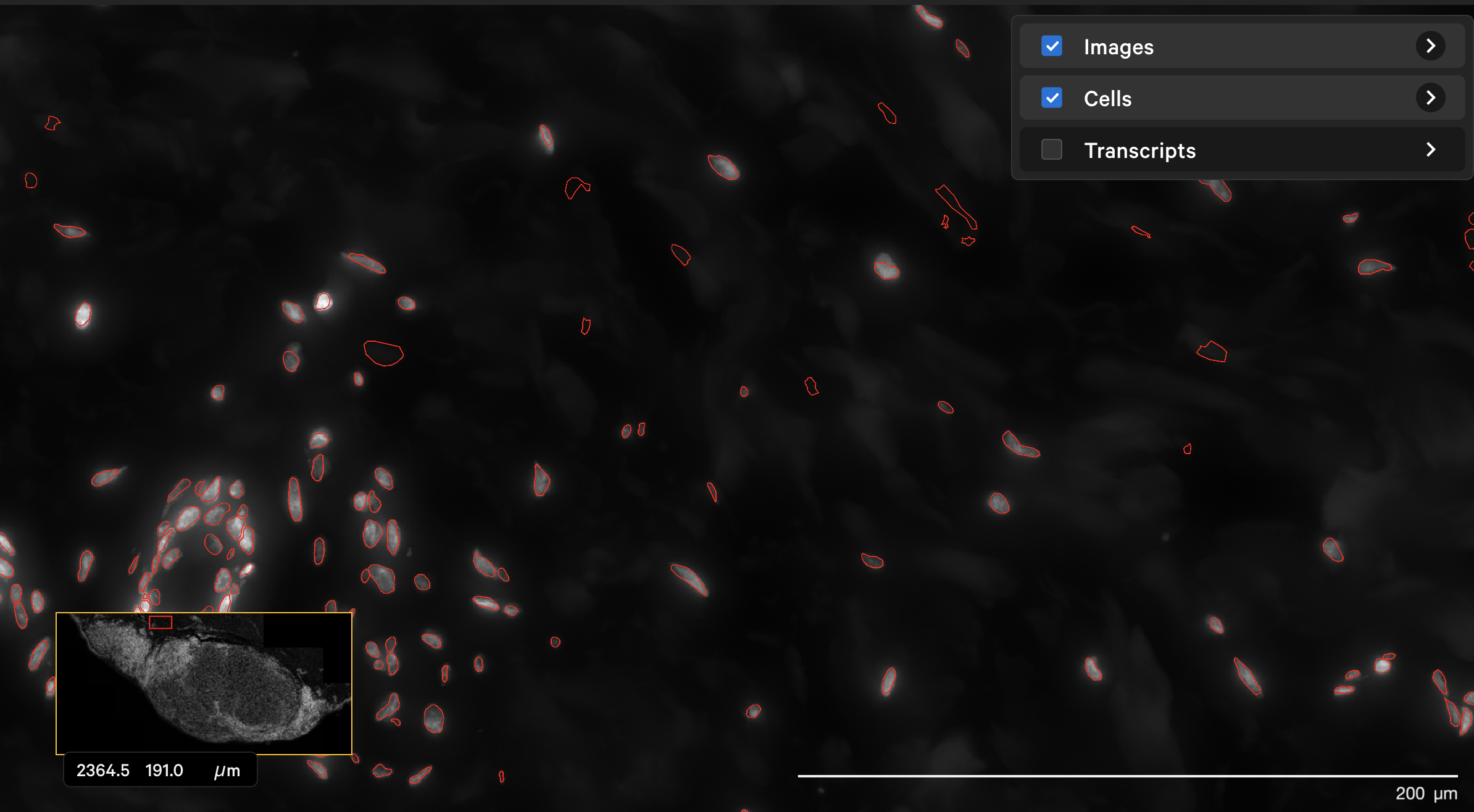
The dapi-filter parameter can also be used to reduce false positives in cell calling. For example, the onboard analysis pipeline identified cells in the dermis in this human skin dataset that are not likely to be true cells. The original DAPI image shown with outlined nuclei in Xenium Explorer looked like this:
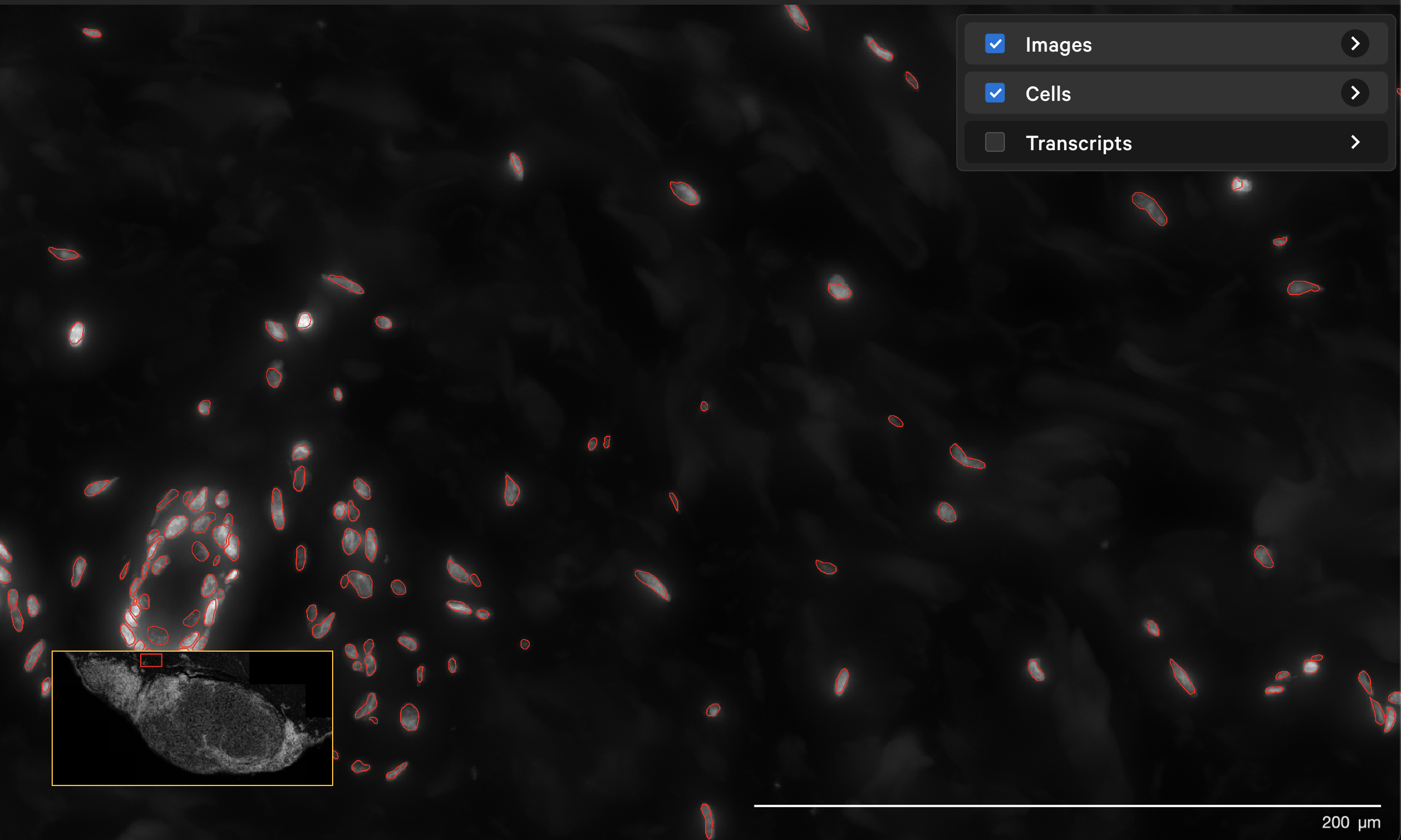
The resegment result with dapi-filter=400 for the same area now looks like this:
After the pipeline completes, check the output outs/ directory (see Output overview). For example, look at these files:
- Analysis summary: check metrics and plots on the Cell Segmentation tab
- All secondary analysis files
- All cell and transcript files
Morphology images are unchanged and nuclei boundaries only change if you are using --resegment-nuclei=true. The updated segmentation results can be visualized in Xenium Explorer.