A journey to new worlds of discovery at AGBT 2023
AGBT is always one of our favorite conferences, and we were particularly excited to take part this year: not just so we could showcase how we’re bringing the latest breakthroughs to your lab, but how these technologies and applications are reshaping what’s possible at the single cell level—and beyond.
Flexing our strength in the single cell revolution
Single cell analyses have revolutionized both basic and translational research. But there’s still so much more it can help you achieve. That was the philosophy behind our Chromium Single Cell Gene Expression Flex, which was developed, in no small part, because single cell options for fixed samples—critical for clinical and multi-site studies—were sorely lacking. This assay enables you to use more sample types—fresh, frozen, or FFPE tissues, as well as fresh or fixed cell or nuclei suspensions—at greater scale, with up to 128 samples and 1 million cells per chip.
In our workshop at AGBT, we showed how this workflow not only offers higher sensitivity and robustness than our Chromium Single Cell 3’ assay, but also incorporates more stopping points, making it even easier to use. On the assay’s simplicity, Jill Herschleb, PhD, Senior Director of Cell Biology at 10x Genomics, offered this proof of principle:
"A single scientist who has never touched lung tissue before [...] followed our protocol with no optimization, modification, sample pre-screening, or qualification. These were just purchased from a biobank. And the experiment was done once. [...] I'd like to emphasize this is a workflow that anyone can run, making analysis of multiple samples a reality. And here are the data from all 32 samples on one Chromium chip, totaling 900,544 cells."
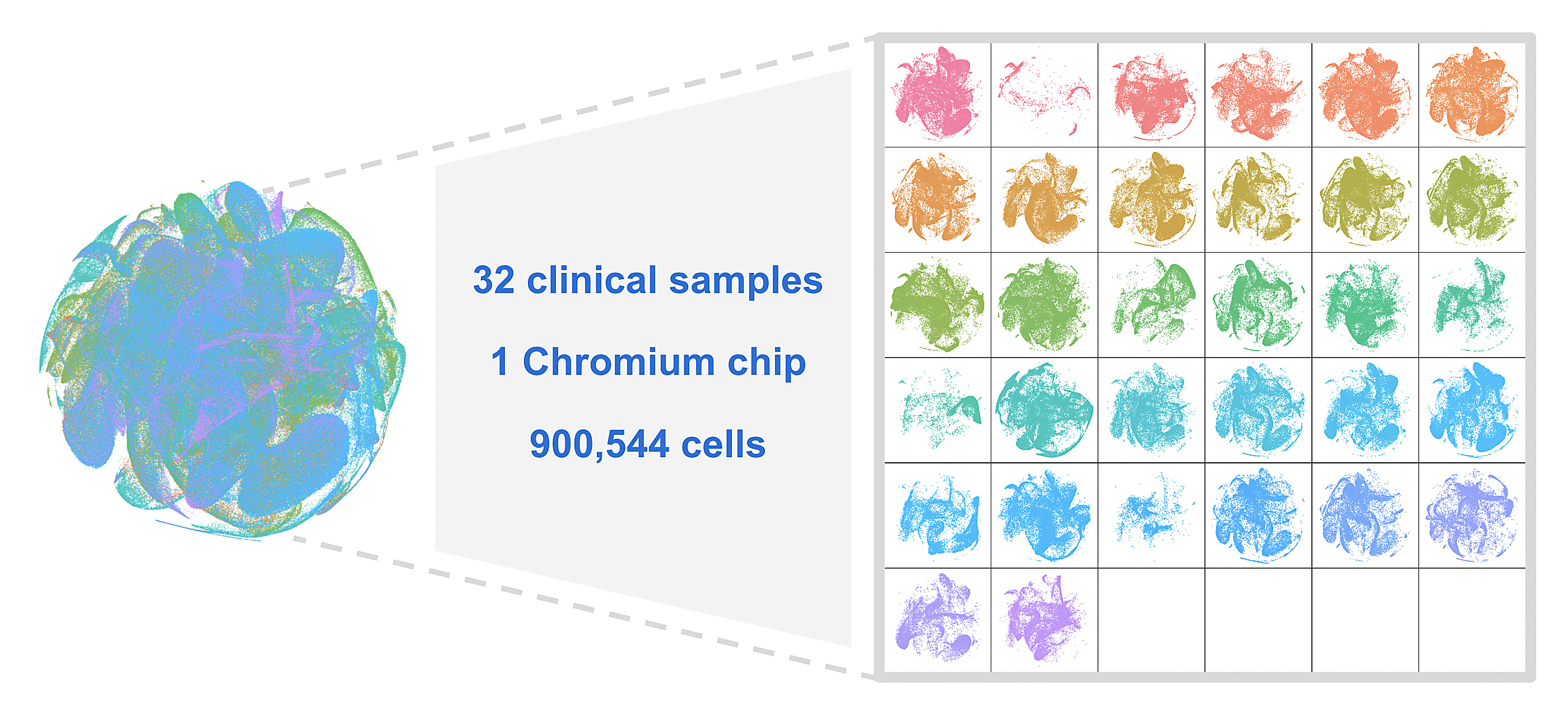
Recapping our validation strategies, we showcased how fixed RNA profiling with Flex retains sample biology, making batching and multiplexing trivial for large, multi-site studies. Last, but certainly not least, we teased preliminary data, showing an experiment in which our R&D team barcoded 8 million cells in only 6 minutes.
Imagine what you could do with that kind of flexibility.
Seeing into new worlds of spatial biology
The “where” is just as important as the “what” in your gene expression studies. The second portion of our AGBT presentation focused on how seamlessly integrating “where” and “what” is critical for a clearer picture of biology—and how our Visium CytAssist instrument can help you get there.
We introduced the Visium CytAssist instrument last year with the goal of expanding sample access and maximizing your insights. We included data that validated this system across dozens of human and mouse FFPE tissues. Later, we revealed the ability to use pre-sectioned fresh frozen tissues with the Visium CytAssist and illustrated how this instrument outperforms direct placement in manual spatial transcriptomics workflows for both FFPE and fresh frozen tissues.
Our presentation highlighted how the CytAssist can give you greater insights with expanded analytes and data integration: one of our AGBT posters showcased the compatibility of the Visium platform with tissue microarrays, and we’re currently finalizing the ability to get morphological, transcriptomic, and protein data from a single slide (coming in the first half of 2023). Finally, we touched on the incredible resolution capabilities of the future Visium HD assay, which was able to resolve extremely fine morphological details in a section of mouse intestine.
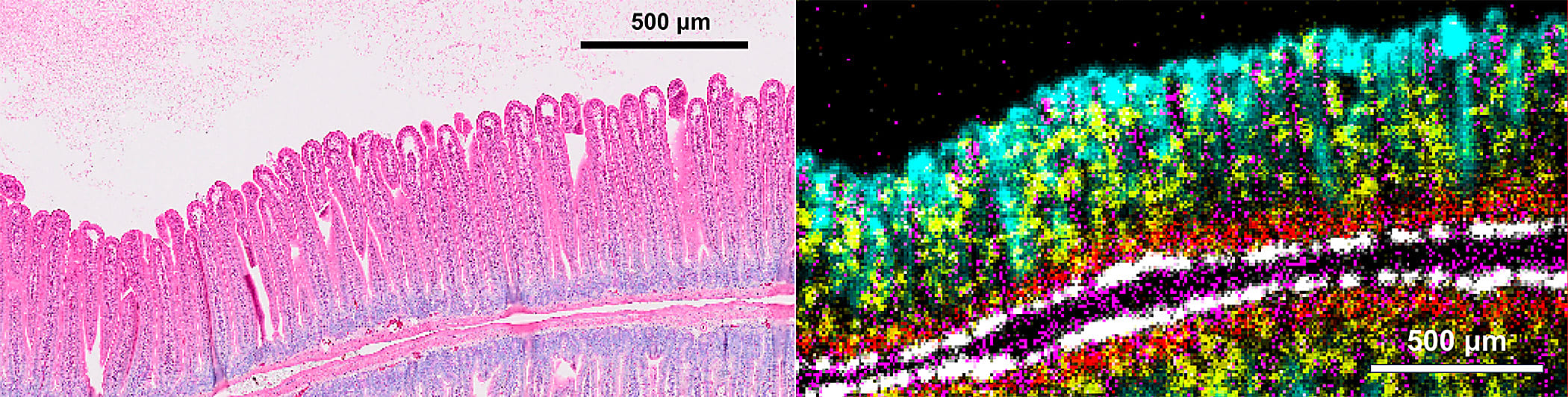
"...And you can see here our single gene markers for the cell types that we would expect in the villi and the crypts of the mouse small intestine. These are not immunofluorescence images, they are NGS-based readouts. And you can see they were able to resolve the fine structures and cell layers in this complex tissue."
- Augusto Tentori, PhD, R&D Lead for Visium
And, speaking of resolution…
Catalyzing research at subcellular resolution
Much like the black box of bulk sequencing, single cell sequencing has a fundamental limit of resolution: a single cell. But transcriptional localization, cell-specific isoforms, and other phenomena may be restricted to certain components of the cell. So, how can we tell what’s really going on in there?
Our answer to that question is the Xenium In Situ Analyzer, a new instrument that is capable of high-throughput, subcellular mapping of 100s to 1,000s of RNA transcripts simultaneously. Our team was excited to show off the instrument’s capabilities to industry, clinical, and basic research leaders—particularly the high sensitivity (it mapped ~280 transcripts per cell in a section of human brain in one experiment) and the fact that its gentle workflow allows for morphological or immunofluorescent staining in addition to the standard workflow. And if transcriptomics aren’t enough for you, you’ll be happy to know that multiomic RNA + protein assays, single nucleotide variant (SNV), and transcriptomic isoform capabilities are part of the planned functionality rollout.
Just as exciting, though, is its future, so we were proud to cap off our talk with the Xenium Catalyst program. We believe the more accessible a technology is, the greater its impact. This program will help you obtain proof-of-concept data for grant applications, help you learn why Xenium is right for you, and more. We’ll have all the information on our website soon, so stay tuned!
Delivering on our promises
Last summer at AGBT 2022, several of these technologies and their possibilities—most notably FFPE compatibility with Single Cell Gene Expression Flex, Visium CytAssist, and the Xenium platform—were only promises. Now, they represent real opportunities for your lab to push your research further than ever before. We were proud to show off some of the posters and customer publications that have already come from these technologies, including:
- Bartosovic M & Castelo-Branco G. Multimodal chromatin profiling using nanobody-based single-cell CUT&Tag. Nat Biotechnol (2022). doi: 10.1038/s41587-022-01535-4
- Guilliams M, et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell 185(2): 379–396 (2022). doi: 10.1016/j.cell.2021.12.018.
- Chromium Fixed RNA Profiling reveals single cell data with comparable biological complexity from FFPE blocks and their matching fresh tissues (Poster 507)
- Evaluating Alzheimer’s disease-associated gene markers with Xenium in situ gene expression (Poster 513)
- And more!
Watch (or rewatch) our workshop here to check out the full spectrum of innovation from us at AGBT this year, and we can’t wait to see you next year!
