Reference-Quality genome assemblies for conservation biology
Dr. Steve Jones and his team at Canada’s Michael Smith Genome Sciences Centre (GSC) used 10x Genomics’ Linked-Read sequencing and Supernova assembler to construct the first publicly available reference genomes for the Beluga Whale (Delphinapterus leucas) and the Northern Sea Otter (Enhydra lutris kenyoni). The final results were published in the December 2017 issue of Genes, and the annotated genome assemblies are available on NCBI.
These assemblies are prized tools for conservation biologists seeking to understand and protect Canada’s dwindling populations of Northern sea otters and Beluga whales. The objectively adorable arctic sea mammals are designated at-risk by the Committee on the Status of Endangered Wildlife in Canada (COSEWIC). Their genome assemblies, however, provide a platform for future conservation efforts, including genetically-informed breeding strategies or the identification of genetic factors that could impart disease resistance in rapidly changing arctic habitats.
I spoke with Dr. Jones last week to find out how his research team, which specializes in cancer genomics, came to lead the effort to construct reference genomes for these two non-model species. As he explains it, the journey began in late 2016 when he saw a flurry of local news stories about Aurora and Qila, a mother-daughter pair of Beluga whales who had fallen ill and then quickly died at the Vancouver Aquarium.
Aurora and Qila had been cared for at the aquarium for over 3 decades, and as the dedicated team of caregivers were mourning the whales’ deaths, they were also scrambling to identify the cause. The sudden demise of two healthy animals was evidence of a possible infection, which could put other animals in their care at risk. Jones knew that a genomic approach could help determine whether a known or new pathogen was to blame for the deaths. Feeling compelled to help solve this urgent mystery, he emailed his ideas to Dr. Martin Haulena, the aquarium's chief veterinarian, who was eager to engage in the proposed collaboration.
With funding from Genome BC and using samples collected by Dr. Haulena’s team, Jones and his group at GSC quickly generated standard short-read genomic and cDNA sequencing libraries from a variety of blood and tissue samples. They then analyzed the resulting sequence data for evidence of infection. Ultimately, they found no evidence of infection in any of the samples tested, which was corroborated by results from a parallel study by the department of Fisheries and Oceans. Based on the combined findings, the cause of death was ruled a toxin, though the exact nature and source remain a mystery.
Dr. Jones then decided to turn the remaining resources, which included both funding and plenty of Beluga gDNA in his lab freezer, towards assembling a reference genome, a resource he knew could play a valuable role in the protection and recovery of Canada’s Beluga populations. To add to their existing short-read genomic sequence data, which they knew lacked the long-range information required to construct a reference-quality assembly, the team constructed and sequenced a single Linked-Read library using Chromium Genome reagents and an Illumina sequencing platform. The raw FASTQ files were loaded into Supernova (v1.1.5), a push-button de novo assembler that provides fast processing and accurate assembly. The resulting Supernova assembly was further improved using the standard short-read data for scaffolding and gap filling. In their discussion, the authors note that their scaffolding workflow provided only a modest gain in assembly quality over what was achieved with Linked-Reads and Supernova alone (Table 1).
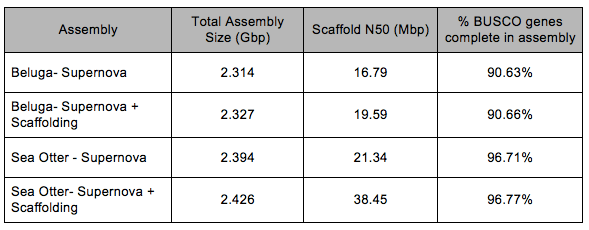
While working on the Beluga project, Jones learned of another genomic study underway at the Vancouver Aquarium. In April 2017, Elfin, a 16-year-old Northern sea otter who had lived at the aquarium since he was rescued as an orphaned pup, died peacefully after a year-long battle with lymphoma. Unfortunately, cancers are not uncommon in marine mammals. Dr. Haulena and his team were now trying to understand the genetic underpinnings of Elfin’s lymphoma with the hope of designing more effective treatment strategies for the future.
But in order to identify genomic changes that may have lead to Elfin’s lymphoma, they first needed a reference assembly to provide the baseline for a healthy geome. Using healthy tissue samples from Elfin and the same assembly workflow they used for Beluga, Dr. Jones and his team were able to quickly generate a reference genome for the Northern sea otter (Table 1).
Following their successes with the Northern Sea Otter and Beluga Whale, Dr. Jones anticipates that his work with CanSeq150, a national genome assembly effort commemorating Canada’s sesquicentennial, will bring more opportunities to collaborate with the Vancouver Aquarium.
Read the full articles for the Beluga Whale and Northern Sea Otter Genome assemblies.
Read more about Dr. Jones’ research group at the Michael Smith Genome Sciences Center.
More Resources:
- Read about de novo assembly of the Hawaiian monk seal genome (Yay! Benny is alive & swimming)
- Learn more about the cost-effective Chromium de novo Assembly Solution and Supernova software for everyday de novo assembly
