Understanding cancer gene regulation with single cell chromatin accessibility CRISPR screens
What is the Innovator Series?
The Innovator Blog Series celebrates research conducted by 10x Genomics customers who have demonstrated scientific ingenuity by adapting Chromium Single Cell or Visium Spatial sequencing-based assays. These innovations are distinguished by their originality and potential impact on scientific discovery, including providing access to novel analytes, advancing multiomic analysis techniques, and demonstrating critical applications to human health and disease.
Sequencing tricks and strategic alterations to amplification protocols for ATAC-seq library prep enabled the science behind our next blog. We highlight research from Stanford University and the Chan Zuckerberg Biohub to integrate single cell CRISPR screens with single cell ATAC-seq. This approach allows simultaneous detection of regions of open chromatin in the genome—for insights into transcription factor activity—alongside single guide RNA—for a view of perturbation effects on those same transcription factors. Read on to learn how scientists are using this technique to make new discoveries into the mechanisms of gene regulation in cancer cells.
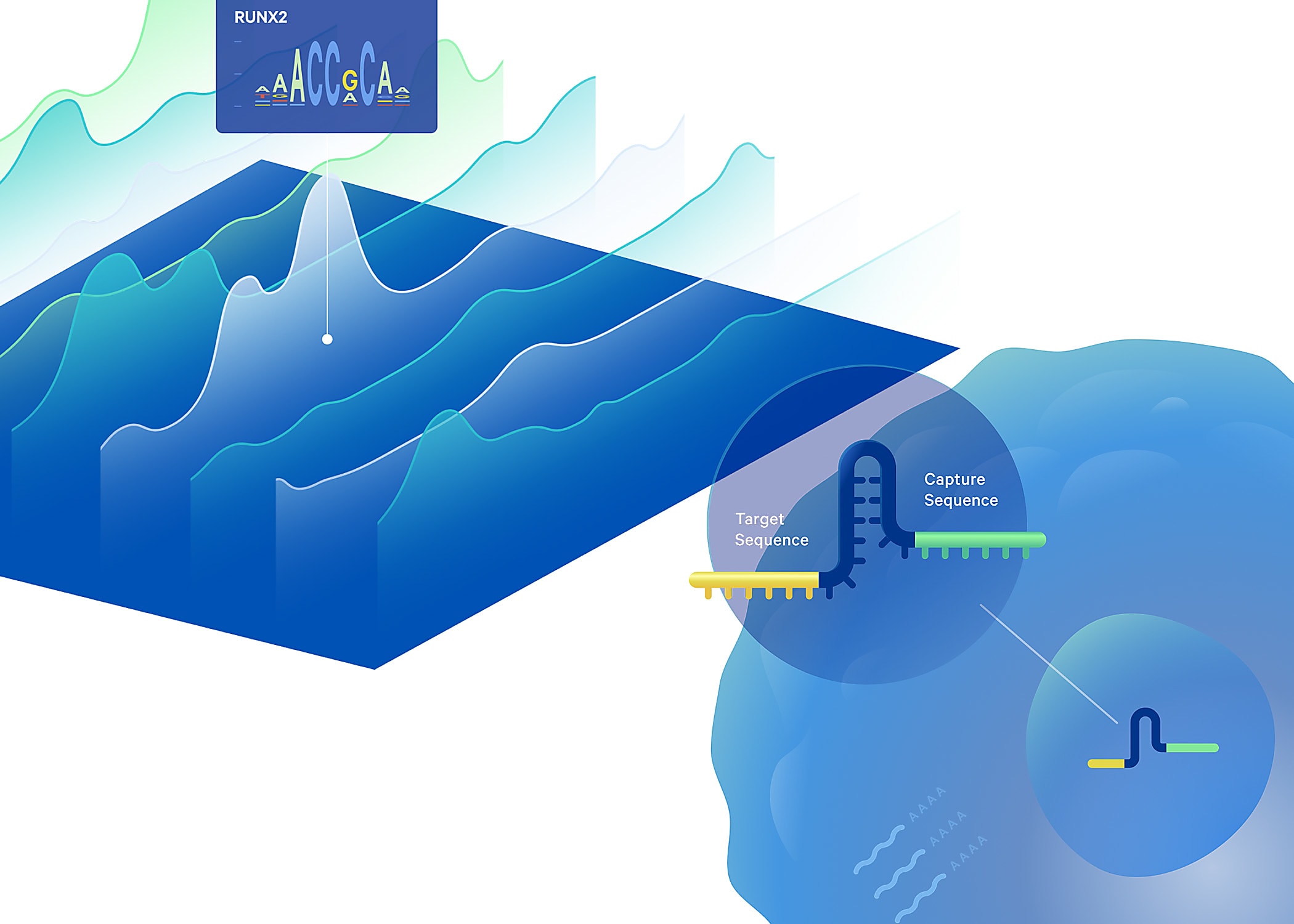
Technology gaps keep gene regulation a mystery
All cells in an individual’s body carry nearly identical genetic information. Despite that commonality, there is incredible biological diversity of cell types and functions, in the contexts of both health and disease. This is a result of unique patterns of gene regulation across different cell types: transcription factors (TFs) and other regulatory elements in the genome interact in precise ways to control gene expression, upregulating the expression of certain genes while downregulating the expression of others. Even cancer cells can find their origin in these unique patterns of gene regulation, though in this case, gene regulation has gone astray, leading to pathology rather than healthy cellular diversity.
Despite the essential nature of these regulatory mechanisms, it remains a significant technical challenge to define the changes TF binding induces in the genome. CRISPR technology has been proposed as a possible solution to determine TF function by perturbing their expression, then tracking the subsequent effects on chromatin accessibility. Though an average readout of TF perturbation on chromatin accessibility across a population of cells could be achieved with bulk screens, gene regulation is highly cell type–specific. Unique patterns of gene regulation could drive cellular heterogeneity even within the same tumor, for example. This makes single cell resolution CRISPR screening an essential capability to understand gene regulation in the context of disease or heterogeneous samples.
This approach also comes with its own challenges, however. While high-throughput single cell CRISPR screens are increasingly effective at perturbing the function of any gene, then observing the resulting effects on the transcriptome, to date, there has been limited development of high-throughput CRISPR solutions that can provide the same readout of perturbation effects on chromatin accessibility, with current methods limited to 96 cells per run (1). This gap in available technology solutions led a team of scientists, including Drs. Sarah Pierce, Jeffrey Granja, and William Greenleaf of Stanford University, to develop a new method called Spear-ATAC (Single-cell perturbations with an accessibility read-out using scATAC-seq), which modifies the 10x Genomics Chromium Single Cell ATAC assay to simultaneously capture single-guide RNA (sgRNA) for a readout of CRISPR perturbation and perturbation effects on chromatin accessibility in the same single cells. With this approach, they were able to assess hundreds of regulatory perturbations in thousands of single cells in parallel, transforming both the quantity and resolution of information that is now accessible in this kind of experiment. Read on to explore the innovations behind their technique and how they’re using it to understand TF activity in cancer.
Tweaking existing protocols to capture sgRNA
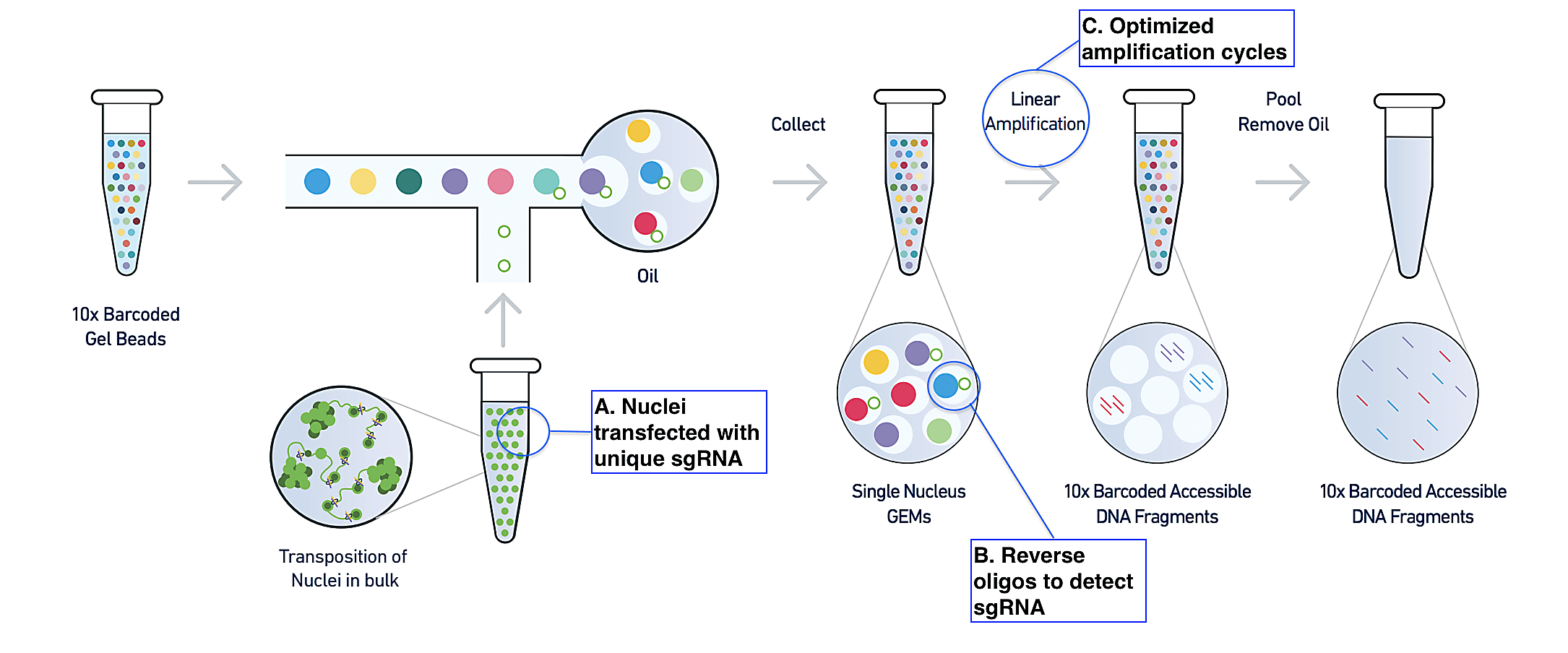
In their recent publication (1) from Nature Communications, Pierce et al. described their novel CRISPR technique. Sharing the same starting point as the standard single cell ATAC-seq assay, Spear-ATAC begins with a suspension of isolated nuclei, which are treated with a transposase (Tn5) enzyme to make cuts in the genome at regions of open chromatin. The method diverges, however, in how nuclei are transfected with sgRNA prior to transposition. To ensure that the assay could detect sgRNA, no matter its accessibility context—in other words, no matter where Tn5 makes cuts in the genome—the research team chose to flank each lentiviral sgRNA with sequencing adapters that would allow amplification of the genomic DNA-integrated sgRNA sequence with the same primers used to amplify ATAC-seq fragments (Figure 1, A).
Following the standard ATAC-seq workflow, the single nuclei suspension is then run through a Chromium instrument where individual nuclei are partitioned into oil droplets with Gel Beads carrying specialized oligos. In these Gel Beads-in-emulsion (GEMs), nuclei content, such as ATAC-seq fragments, are tagged with a nucleus-specific barcode that serves to trace fragments back to the nucleus of origin. Again, to ensure the sgRNA is detected and linked to the nucleus of origin in the same steps, the authors leveraged a reverse oligo specific to the sgRNA backbone. This oligo tags the sgRNA and is then itself tagged with the nucleus-specific barcode, enabling a linked readout of the perturbation and its effects on chromatin accessibility in the same nucleus (Figure 1, B).
A series of changes to the number of amplification cycles in the protocol, and the addition of a biotin-tagged primer to more specifically target sgRNA during these cycles, rounded out the authors’ modifications (Figure 1, C). Taken together, these experimental innovations enabled a 40-fold increase in their ability to detect sgRNA compared to traditional lentiviral integration followed by the single cell ATAC-seq protocol (1). In addition to bridging the technology gap and solving this biological problem of accessing robust chromatin accessibility information in the context of a CRISPR screen, Spear-ATAC also demonstrated significant advantages in cost per cell, time, and scale. For example, while plate-based methods would require multiple iterative experiments over several days to assess just several hundreds of nuclei, in theory, Spear-ATAC could recover up to 80,000 nuclei at once within a 20 minute run on a 10x Genomics Chromium Controller (1).
New insights into GATA1 transcription factor activity in cancer
Though the experimental innovations of this technique are compelling, how the technique translates to scientific discovery is equally important. What kinds of insights into transcription factor activity can Spear-ATAC provide? To address this question, the authors established a test sgRNA library targeting two TFs involved in hematopoietic differentiation and development, GATA1 and GATA2, as well as non-targeting controls, and introduced the library into K562 leukemia cells engineered to express CRISPR-interference dCas9-KRAB, enabling knockdown of the TFs. With data from Spear-ATAC, they developed an analytical framework to identify changes in TF motif accessibility across leukemia cells carrying different sgRNAs. This revealed some expected findings: cells carrying GATA1-targeting sgRNA showed reduced chromatin accessibility at the GATA1 locus, suggesting these cells were downregulating expression at this locus.
The perturbations affected chromatin accessibility at more than these specific regions of the genome, however. Differential accessibility analysis between cells with GATA1-targeting sgRNA and non-targeting controls revealed, overall, 14,262 peaks (14.76%) that increased in accessibility and 14,026 peaks (14.52%) that decreased in accessibility as a result of GATA1 perturbation. Peaks that decreased in accessibility corresponded to areas of the genome near erythroid-specific genes, while genomic regions with increased accessibility following GATA1 knockdown were near megakaryocyte-specific genes. This finding suggests that knocking down GATA1 in K562 leukemia cells may push the cells towards a megakaryocyte lineage. Not only does this position GATA1 as an important factor in this differentiation process, but it could also explain why some patients with genetic disorders that impair GATA1 function show dysregulated production of red blood cells and increased incidence of acute megakaryoblastic leukemia (1).
Expanding the potential of Spear-ATAC
These innovative methods are at their earliest stages of development, with ongoing experimentation revealing greater potential for Spear-ATAC. Looking to push the throughput limits of the method, the Stanford team subsequently established a CRISPR library to knock down 36 transcription factors in leukemia cells, with 2–3 sgRNAs targeting each TF, plus 14 control non-targeting sgRNAs and 12 sgRNAs targeting essential genes. In this scaled experiment, they captured 32,832 nuclei from six samples, covering 128 different sgRNA genotypes (1). With high-throughput capabilities and high-resolution insights across many different perturbations, the research team anticipates exciting applications for Spear-ATAC, such as generating TF interaction maps that reveal how proteins interact to regulate the non-coding genome, and even how mutations in the context of disease can affect TF accessibility motifs.
Incredible technological innovation has brought these complex biological questions and their answers within reach. Making the impossible possible is at the heart of innovation; and, thus, Spear-ATAC represents a very worthy member of the growing list of technologies that make up our Innovator Series.
Learn more about Spear-ATAC here →
References:
- Pierce S, et al. High-throughput single-cell chromatin accessibility CRISPR screens enable unbiased identification of regulatory networks in cancer. Nat Commun 12: 2969 (2021). doi: 10.1038/s41467-021-23213
