Single cell RNA-seq elucidates the mechanism for a combination therapy in a subpopulation of metastatic colon cancer
Impact at a glance: A Phase 2 clinical trial leveraged single cell RNA-sequencing (scRNA-seq) to study the efficacy of a combination therapy in a group of patients with a mutated, metastatic colon cancer. Single cell data and correlative patient outcomes suggested the therapy was effective to extend progression-free survival in some patients, likely as a result of upregulating immune programs in tumor cells (1).
How can single cell and spatial tools amplify insights in Phase 1–3 clinical trial research? We answer that question in our Clinical trials to watch blog series, featuring innovative applications of single cell multiomics and spatial gene expression to clinical samples. In part one of the series, we showcased a Phase 1 study of an antibody therapy against metastatic endometrial cancer. Keep reading to explore findings from this Phase 2 study.
Phase 2: Targeted therapies cooperate with immune checkpoint blockade
"In what we believe is one of the first clinical trials to incorporate systematic scRNAseq analysis of paired pre- and on-treatment tumor biopsies from all patients, we identified a potential mechanism underlying the cooperativity observed between BRAF/MAPK inhibition and immune response." - Tian J, et al. (Nature Medicine)
Cancer type: Colorectal carcinoma
Therapy: Combined PD-1, BRAF, and MEK inhibition with sparatlizumab (PDR001), dabrafenib, and trametinib, a proof-of-concept single-arm therapy
Trial aim: Determine the safety and efficacy of the therapy, according to the metrics of overall response rate, disease control rate, duration of response, and progression-free survival. Define potential mechanisms of cooperativity between therapeutic agents.
Leading institutions: Massachusetts General Hospital Cancer Center and Harvard Medical School, The Broad Institute of MIT and Harvard, and Gladstone-UCSF Institute of Genomic Immunology
Pharmaceutical collaborators: Novartis Institutes for BioMedical Research
ClinicalTrials.gov registration: NCT03668431
Why was combination therapy tested for this particular cancer type (BRAFV600E colorectal cancer)? The B-Raf proto-oncogene (BRAF)-V600E mutation is present in multiple cancer types, including melanoma, colorectal and thyroid cancer, and some hematological cancers (2). 8–12% of patients with metastatic colorectal cancer (CRC) have this mutation, which is associated with a poor response to chemotherapy and shorter overall survival (3).
While 60-80% of patients with BRAFV600E melanoma respond to BRAF inhibitor monotherapies (including vemurafenib and dabrafenib), less than 5% of patients with CRC tumors harboring the same mutation do (1). To increase BRAF inhibitor efficacy in patients with CRC tumors, researchers targeted components of the mitogen-activated protein kinase (MAPK) signaling pathway—which is constitutively activated in BRAFV600E tumors, promoting uncontrolled growth and apoptotic resistance. Prior research—including studies led by the investigators of this Phase 2 clinical trial—revealed MAPK signaling was reactivated after BRAF inhibition in patients with resistant CRC tumors (1). The FDA recently approved a combination therapy targeting both pathways—BRAFi encorafenib, plus the anti-EGFR antibody cetuximab—in patients with BRAFV600E CRC. However, this combination therapy is still only effective in 20% of patients, and its effects only last a few months.
However, a subset of BRAFV600E CRC patients with a particular tumor feature called microsatellite instability (MSI), caused by DNA mismatch repair deficiency, had more durable responses to BRAF/MAPK pathway inhibition. Since patients also respond better to immune checkpoint blockade (ICB) therapy than those with metastatic microsatellite stable (MSS) CRC, the investigators hypothesized that BRAF/MAPK inhibition may enhance the immune response in metastatic BRAFV600E CRC (1). Thus, this combination of targeted therapies with an immune checkpoint inhibitor, which enhances patient anti-tumor immune responses, has the potential to address unmet clinical needs for varying subgroups of BRAFV600E colorectal cancer patients.
Clinical results: 37 patients (including 32 metastatic CRC patients with MSS, and 5 metastatic CRC patients with microsatellite instability (MSI), an ICB-responsive subclass of CRC) were treated with the combination therapy (sparatlizumab, dabrafenib, and trametinib) for a median of 7.4 months. 9 patients (24.3%) showed a confirmed response, including 25% of MSS patients, which is a favorable outcome compared to previous clinical studies of dabrafenib and trametinib alone in BRAFV600E MSS CRC patients (previously, a 7% confirmed response rate (1)). Two patients had a complete response. The trial influenced progression-free survival and overall survival rates, with a median of 4.3 months and 13.6 months, respectively (1).
Of the 32 MSS patients, those who were predicted to have an insignificant response to ICB alone, 28 had no prior treatment with BRAF inhibitors. These patients saw a median progression free survival of 5 months, with 5 patients staying on the therapy for over a year. One of these patients showed a partial response over the next 2.5 years (1).
Applications of single cell sequencing: Given these positive results relative to monotherapy regimens or combination therapies without ICB, the research team sought to understand the basis of therapeutic response—in particular, noting the cellular and transcriptional effects of BRAF/MAPK pathway inhibition—and if there was significant enhancement of tumor-immune responses. Scientists took core needle biopsies from all patients before treatment and on day 15 following treatment from the same lesion. They were ultimately able to conduct single cell analysis on samples from 23 patients for a total of 419,551 single cells.
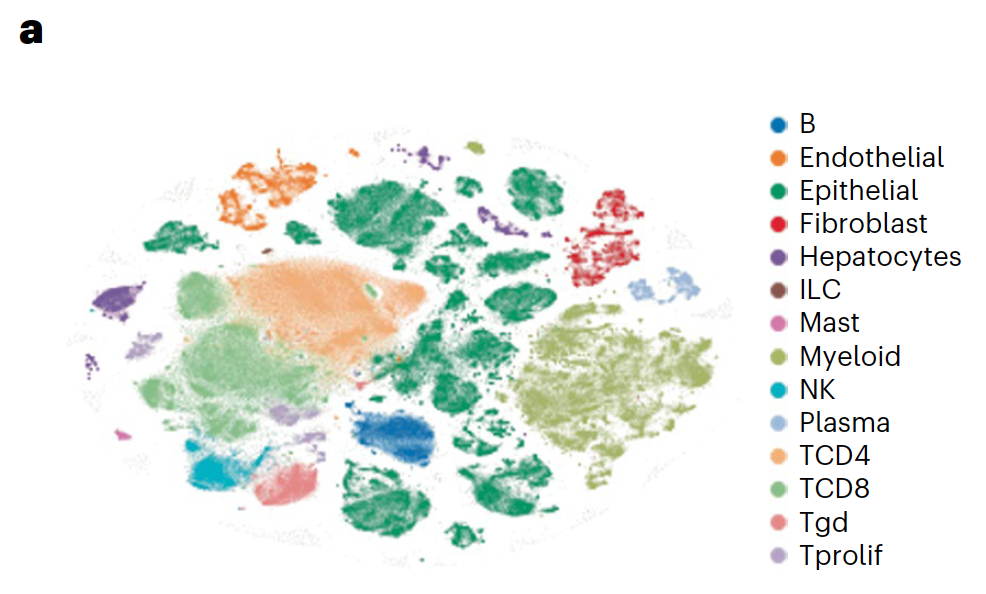
Using Chromium Single Cell 5’ Gene Expression, the researchers performed single cell RNA-seq to observe changes in cellular composition between pre- and on-treatment biopsies. In patients who experienced progression free survival (PFS) over 6 months, they observed a significant decrease of tumor epithelial cells and an increase of CD45+ immune cells and CD8+ T cells (1). The team also observed a spike in expression of immune-related genes—such as genes involved in the interferon response, antigen processing and presentation, and chemokine activity—in tumor cells from patients with over 6 months PFS, but not in tumor cells from patients with less than 6 months PFS (1).
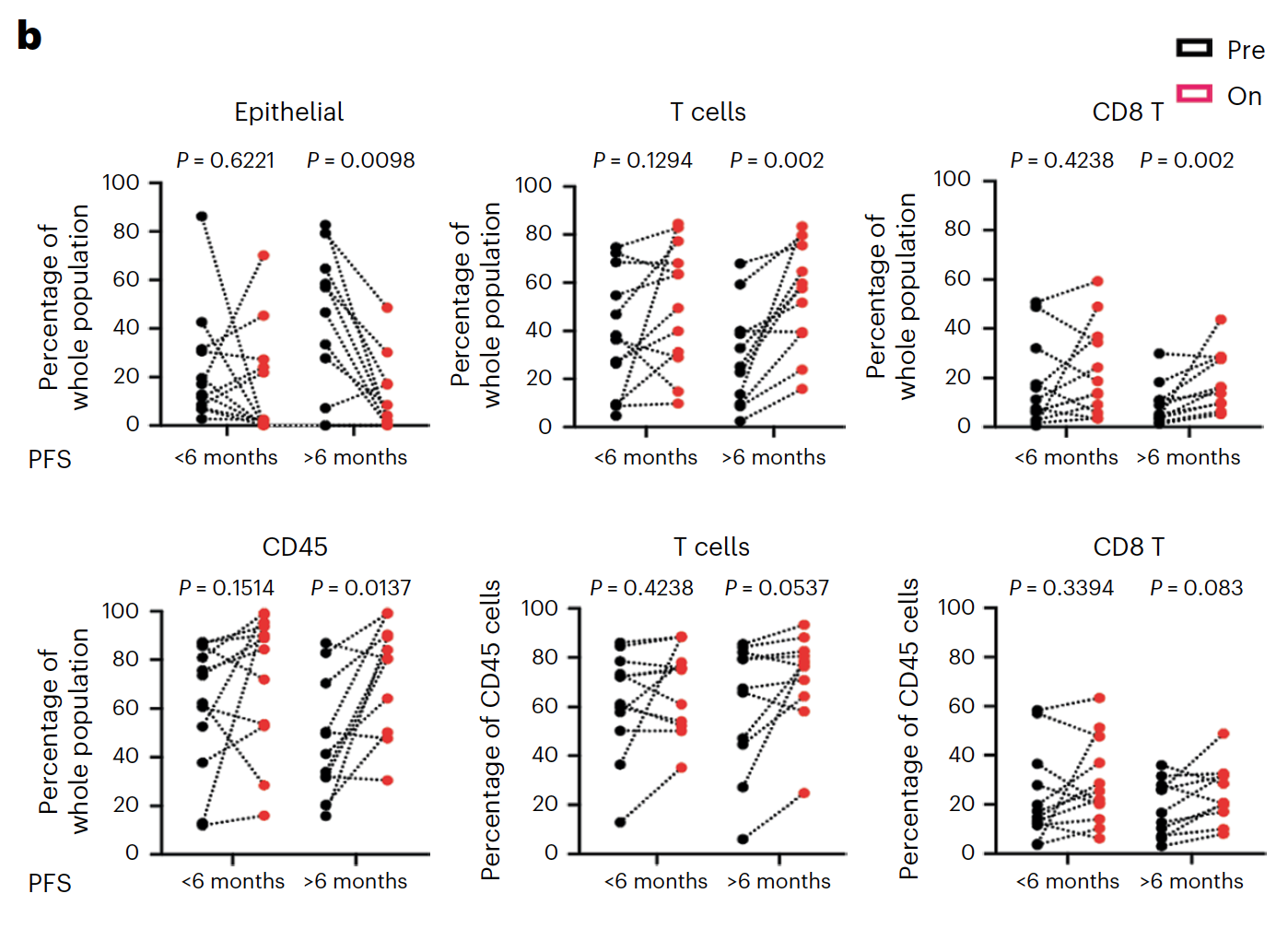
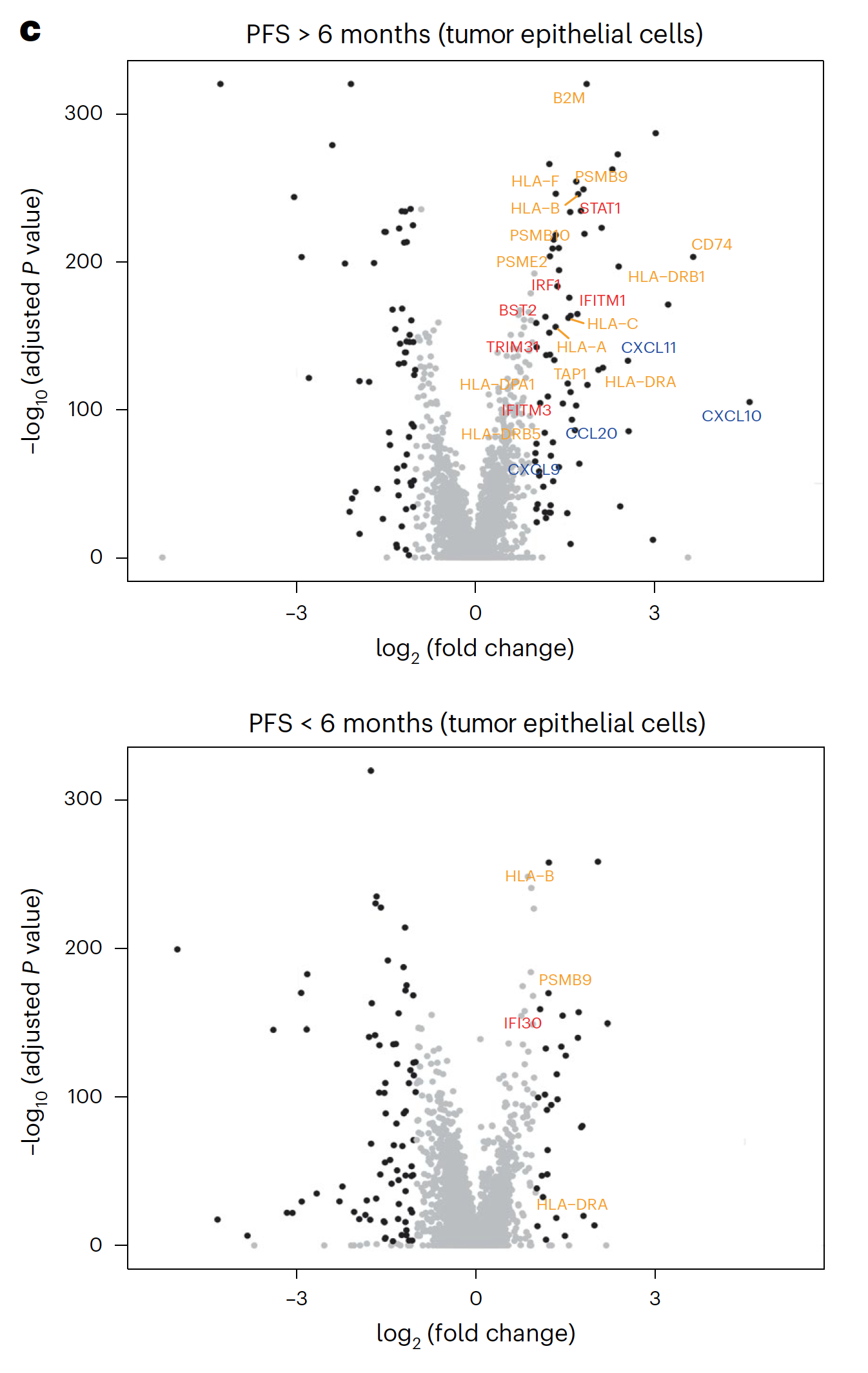
These single cell findings ultimately point to a possible mechanism of cooperative activity underlying the triple combination therapy with sparatlizumab (PDR001), dabrafenib, and trametinib in BRAFV600E colorectal cancer. BRAF/MAPK pathway inhibition may upregulate immune gene programs in tumor cells, including chemokines that serve to recruit activated T cells, providing the therapeutic benefits of a stronger anti-tumor immune response as seen in patients with longer PFS. The study authors suggest this may be a critical mechanism underlying therapeutic efficacy (1).
Understanding the cellular and molecular differences between patients with diverging clinical outcomes using high-resolution single cell insights therefore represents a robust approach to interrogate therapeutic mechanism of action—and, in this trial, an important step to determine if combination therapies can provide clinical benefits to not only BRAFV600E CRC, but to other cancer types with the same affected signaling pathways.
Read the full clinical trial publication.
Learn more about the technology that supported these clinical findings:
- Chromium Single Cell Immune Profiling (5’ Gene Expression)
Download our White Paper to learn how single cell tools can overcome barriers to drug discovery and development.
References:
- Tian J, et al. Combined PD-1, BRAF and MEK inhibition in BRAFV600E colorectal cancer: a phase 2 trial. Nat Med 29:458–466 (2023). doi: 10.1038/s41591-022-02181-8
- Loo E, et al. BRAF V600E mutation across multiple tumor types: Correlation between DNA-based sequencing and mutation-specific immunohistochemistry. Appl Immunohistochem Mol Morphol 26:709–713 (2018). doi: 10.1097/PAI.0000000000000516
- Tabernero J, Ros J, and Élez E. The evolving treatment landscape in BRAF-V600E–mutated metastatic colorectal cancer. Am Soc Clin Oncol Educ Book 42:1–10 (2022). doi: 10.1200/EDBK_349561
