Untangling immune complexity in SARS-CoV-2, HIV, and pneumonia: Highlights from the Global Infectious Disease Symposium
At this point—nearly two years into the COVID-19 pandemic—we all understand the impact of infectious disease and why research in this field is so critical. However, the study of infectious disease comes with unique challenges. The immune response to disease is incredibly diverse, with each and every step branching into complex mechanisms and cellular interactions. And while immunologists work to decipher this vast network, they must also find ways to keep up with variants, mutations, and novel/emerging pathogens. Overcoming the challenges of infectious disease requires a deeper understanding of the fundamentals of these complex pathologies, from the development of innate and adaptive immunity to the intricacy of host–pathogen interactions and immune evasion. But don’t just take our word for it.
At this year’s Global Infectious Disease Symposium, leading scientists shared their latest findings, giving us an inside look at how they’re using innovative single cell technologies to advance our understanding of the immune response to infection. Keep reading to find out more about their infectious disease research. Then, watch the full presentations on-demand.
Deconstructing the form and function of T cell responses to infections
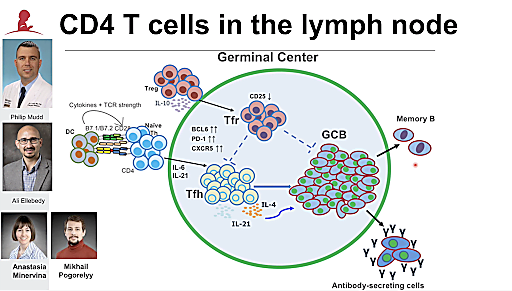
Early in the COVID-19 pandemic, Dr. Paul Thomas and his colleagues at St. Jude’s Hospital began to build a cohort of samples tracking the immune response to SARS-CoV-2 and, later, vaccination. Dr. Thomas, of course, knew the value of this information; the primary focus of his lab’s research is T-cell specificity and how it relates to T-cell function and response efficacy when faced with infection.
First, Dr. Thomas and his team divided the cohort into four groups—patients who had been infected by SARS-CoV-2, those vaccinated against SARS-CoV-2, those who were vaccinated after infection, and those who were infected after vaccination, or breakthrough cases. Hypothesizing that the magnitude and specificity of the T-cell response against vaccines and pathogens is a critical determinant of response efficiency, the team used a combination of single cell gene expression, single cell immune profiling, and a technique Dr. Thomas calls “reverse immunology” to investigate immune response in individuals within the four groups. They found significant differences in response among the groups. For example, breakthrough cases did not display the enrichment of spike protein–specific antibodies that is associated with a robust immune response to SARS-CoV-2. This suggests that in these cases, despite priming, the immune system is not effectively recalling the spike-specific response.
Dr. Thomas also shared some of the computational strategies that he and his team used to delve deeper into antigen specificity. After paired T-cell receptor (TCR) profiling, the researchers used an algorithm called TCRdist, which allowed them to look, collectively, at groups of TCRs and determine their relationships to one another. Using this technique, they were able to identify major motifs in the SARS-CoV-2 response, and found that vaccination and infection elicit equivalent memory repertoires, pointing to the efficacy of the vaccine and its ability to mirror infection in recruitment and selection of TCRs into the repertoire.
Learn more about Dr. Thomas’s research by watching the full on-demand webinar →
The local and systemic response to SARS-CoV-2 infection in children
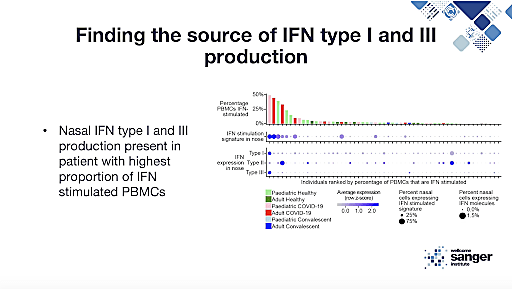
We got a different perspective on the COVID-19 pandemic with Dr. Rik Lindeboom, a member of Sarah Teichmann’s lab at the Wellcome Trust Sanger Institute, whose research focuses on response to SARS-CoV-2 in children. Though we know that children respond very differently to SARS-CoV-2 infection than adults, usually displaying mild symptoms or, often, none at all, the cellular and molecular mechanisms underlying this difference are not well understood. However, as Dr. Lindeboom explained in his presentation, knowing what causes this variation in response could greatly inform our understanding of the disease, especially the factors that drive severity.
Studying a cohort of pediatric and adult patients, a mix of infected and healthy individuals, Dr. Lindeboom and his colleagues collected four samples from each patient—nasal, tracheal, and bronchial brushes, as well as matched PBMC samples. They then used a combination of techniques, including single cell gene expression and immune profiling, to generate a cell atlas of the upper airway according to age and over the course of COVID-19. Using the atlas, they were able to compare SARS-CoV-2 response in both the airway and the blood. With this local and systemic data, ranging both age and stage of disease, the team uncovered insights into maturation patterns of the immune system and how they affect immune response. Further, they were able to isolate the pediatric-specific response, uncovering crucial information about mechanisms within the pediatric immune system that are protective against infection and how these findings may inform future therapies.
Find out more in Dr. Lindeboom’s full on-demand webinar →
Novel Insights into Lung Immunity: From pneumonia to COVID-19
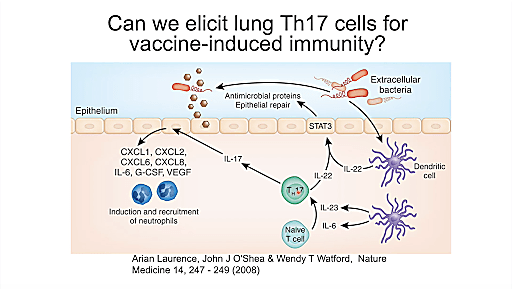
Next up Dr. Jay Kolls, Director of the Center for Translational Research in Infection and Inflammation at Tulane Medical School, gave us an inside look at his investigations of lung immunity in both pneumonia and COVID-19. Using single cell RNA-sequencing (scRNA-seq), Dr. Kolls sought to understand the cellular relationships in the context of pneumonia, particularly within the dynamics of host defense mechanisms. Focusing on Klebsiella pneumoniae, a hospital-acquired bacterial infection that has developed multiple drug-resistant strains, Dr. Kolls used knockout mice to understand the factors driving susceptibility and was able to observe mechanisms of the adaptive immune response that have the potential to inform future vaccination strategies. One possible application, for example, is the development of inhaled vaccines.
Switching gears, Dr. Kolls then walked us through his study of SARS-CoV-2 infection in transgenic mice expressing ACE2, a receptor for the virus. Four days after infection—the start of lung pathology—Dr. Kolls and his colleagues used scRNA-seq to evaluate the transgenic mice as well as non-transgenic control mice also infected with SARS-CoV-2. Clustering the cells, they found that transgenic mice displayed a massive amount of inflammatory monocytes, fibroblast activation and proliferation, and an increase in T cells. Finding viral RNA in all of these clusters in the transgenic mice but not in control mice, they used Loupe Browser, our visualization software, to re-cluster the virus-positive cells, leading them to discover a cluster of viral cells that they hypothesize may be exosomes. Dr. Kolls’s next order of business will be to uncover where the viral RNA first traffics, hoping to better understand SARS-CoV-2 pathogenesis. He expects it’s epithelial cells, but, like you, we’ll have to wait to find out.
Watch the full on-demand webinar to learn more about Dr. Kolls’s findings →
HIV and the brain – A single cell analysis of initial infection, its maintenance, and the effect of treatment
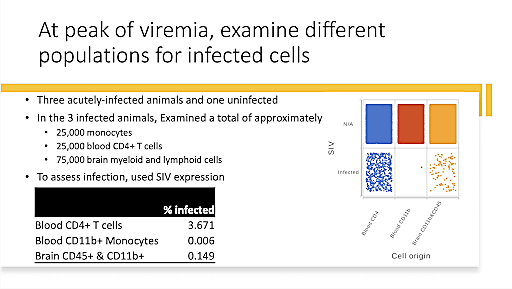
Dr. Howard Fox, University of Nebraska Medical Center, ended the Global Infectious Disease Symposium on a high note, sharing his research on how HIV infection affects the brain. Early in the infection, the virus enters the brain, causing some HIV patients to develop cognitive disorders, such as encephalitis and dementia, a phenomenon that Dr. Fox calls neuroHIV. Though people living with HIV can now survive to a near-normal lifespan with treatment, the chronic nature of the infection means that cognitive dysfunction and disorder is still a possibility as the illness persists over decades. Further, it is not well understood how HIV affects or interacts with age-related diseases like neurodegenerative disorders. There is also a high rate of substance abuse in people living with HIV, another factor with unknown effects on neuroHIV.
Using single cell technologies to study SIV, an infection very closely related to HIV, in non-human primates, Dr. Fox aims to understand how HIV infection of macrophages and microglia affects the central nervous system (CNS), causing brain dysfunction. He began by developing a strategy to overcome the challenges of microglia isolation, allowing the cells to be cryopreserved while maintaining critical disease-related gene expression patterns. This allowed him to move forward in studying neuroHIV microglia in various conditions, including methamphetamine and opioid administration. Then, Dr. Fox shifted his focus to cell trafficking and viral persistence in the brain, factors that can act as a barrier to a cure for HIV. Using the Chromium Single Cell Multiome ATAC + Gene Expression assay, he was able to characterize HIV integration sites, some of which were observed in more than one animal, suggesting that there are preferential integration sites within the brain. Dr. Fox’s research into this fascinating subject is still in progress, and we can’t wait to see what he discovers.
Find out more about Dr. Fox’s study of HIV and the brain in the full on-demand webinar →
