Future releases of Cell Ranger will not support Low Throughput (LT) libraries.
The Chromium Next GEM Single Cell 3’ LT v3.1 kit (referred to as low throughput or LT) is a low throughput, cost-effective solution for smaller-scale and pilot studies for profiling whole transcriptome at the single cell level for 100 - 1,000 cells per sample. In combination with Feature Barcode technology (Antibody Capture), the assay also enables simultaneous cell surface protein detection in single cells.
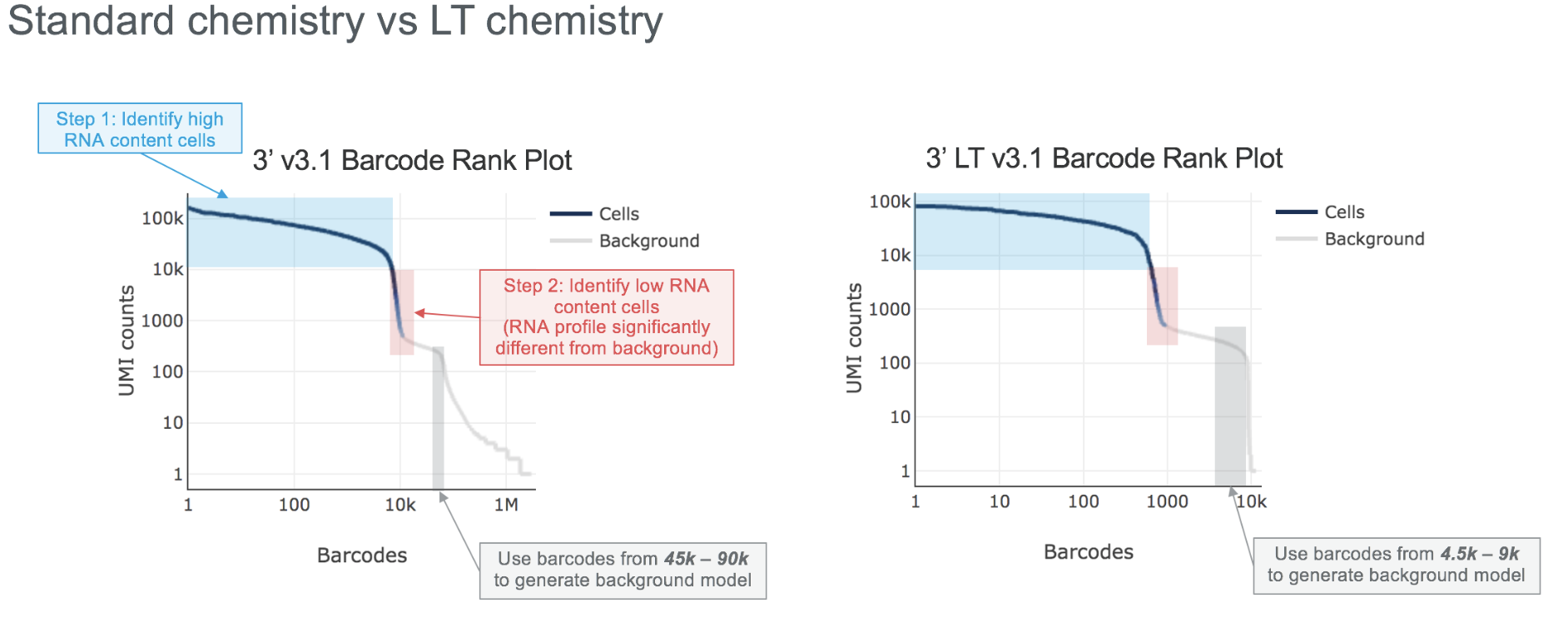
Cell Ranger v6.0 or later is required to analyze LT data. After demultiplexing the BCL files with cellranger mkfastq, run the cellranger count pipeline on the FASTQ data to obtain output files. The pipeline will autodetect LT libraries, in which case the chemistry tab in the web_summary.html will show Single Cell 3' v3 LT. One can also use the --chemistry=SC3Pv3LT argument when running cellranger count (this is only recommended if there is an error in automatic detection).
The processing of LT libraries is identical to standard libraries except for minor adjustments to certain parameters in step 2 of the cell calling algorithm for optimal results (see the graphic below for details).
cellranger reanalyze is used to re-run secondary analysis with custom parameters, and cellranger aggr is used to aggregate the outputs of multiple samples, including LT chemistry data. For the latter, one can also aggregate the results of multiple LT libraries, or LT and standard 3' v3.1 libraries, with no chemistry batch correction needed.