InPSYght sequencing project incorporates Linked-Read data
Linked-Reads are a powerful tool for genomic study, transforming the capability of existing short-read sequencers to provide improved interrogation of hard-to-map regions of the genome, superior structural variant detection and haplotype information.
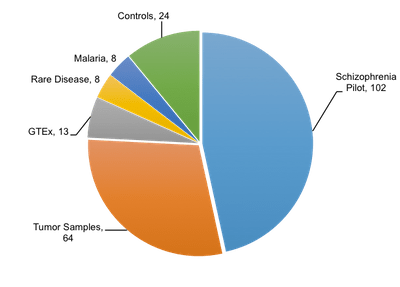
Researchers at the Broad Institute, recognizing the advantages of Linked-Reads, have begun evaluating the Chromium system for integration into their research methods. As part of our pre-symposium workshop at AGBT this year, Stacy Gabriel presented data showing scientists at the Broad have already produced 219 Chromium™ libraries. The pie graph to the right breaks down the variety of ways in which they’ve used the Chromium™ System, so far. As you can see, the biggest slice belongs to the Schizophrenia Pilot project.
The pilot project is part of the much larger InPSYght schizophrenia and bipolar disorder sequencing project, which seeks to produce Whole Genome Sequencing (WGS) data for 10,000 schizophrenia, bipolar, and control samples. Ultimately, researchers hope to use this data to identify variants associated with disease, build a reference panel of structural events and haplotypes to impute into large genotype data sets, and create and share data and methods with the community. After a successful pilot project, the InPSYght data will include WGS data for 500 samples generated using Chromium Linked-Read technology.
In a recent GenomeWeb article, Psychiatric Disorder Sequencing Project Adds 10x Genomics Tech for Phasing, Structural Variant ID, Chris Whelan, Computational Biologist at the Broad Institute McCarroll Lab and the Stanley Center for Psychiatric Research, explains more about InPSYght and how linked-reads have become an important part of the project. Read it here.
The pilot project tested the advantages of linked-read technology in generating WGS data for 50 samples, which was then compared to data from same samples generated using standard Illumina PCR-free WGS. Researchers identified several advantages in using Linked-Reads in generating WGS data.
First, Linked-Reads allow you to interrogate hard-to-map regions of the genome that are normally inaccessible with standard short-read sequencing. In the example below, Broad scientists mapped 322 protein coding genes that were not mapped in the standard data.
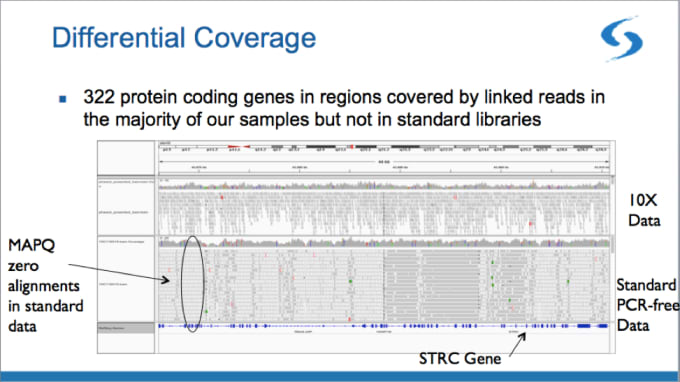
With its barcode-aware alignment, linked-read technology makes it possible to map regions with close paralogs, segmental duplications, and pseudogenes. Anchored by confidently-mapped reads in regions outside hard-to-map areas, the barcodes can then sort short reads into paralogous gene loci.
This barcode-aware alignment facilitates the detection of structural variants (SVs) that are normally undetectable using standard, short-read sequencing. Below is an example of a structural variant that was detected using Linked-Reads. Since the SV spans heterozygous deletion, it is much more difficult to detect using standard, short-read sequencing.
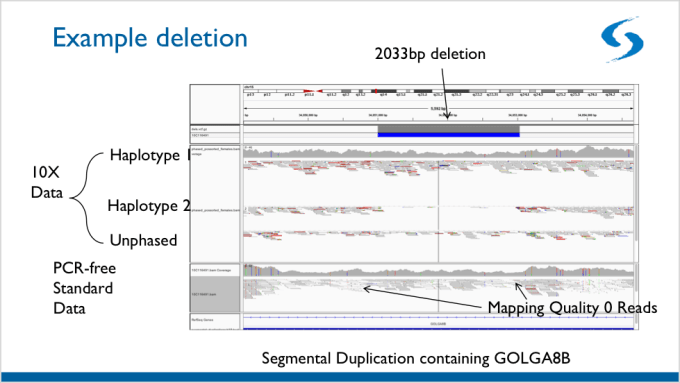
Finally, using linked-reads, researchers were able to identify phased haplotypes for imputation and association, which will allow researchers to more fully examine the rest of the InPSYght data.
You can learn more about the pilot project in our Scientific Seminar video, Exploring 50 whole genomes with Chromium™ Linked-Reads.
Plus, find out more about the advantages of linked-reads in our blog post, Everything you wanted to know about Linked-Reads.
