New resources for enhancing your single cell oncology research
Explore the latest discoveries in cancer biology at our upcoming Global Virtual Cancer Symposium and find valuable resources for your own research. Download our new guide for getting started with single cell applications to see how each step in your experimental workflow plays a part in generating high-quality data and discovering novel biological insights.
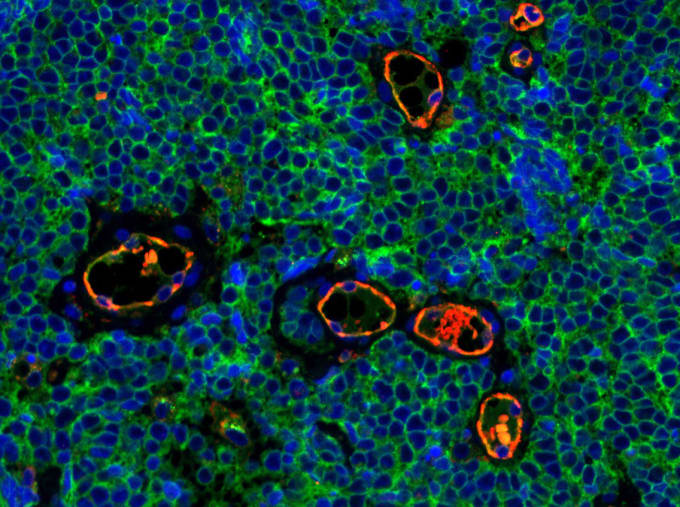
With the ability to more deeply characterize populations cell by cell, scientists are gaining a better understanding of cellular responses to chemotherapy (1), the origins of intratumoral heterogeneity and its relationship to patient survival (2), the mechanism of action for immunotherapy and underlying causes of resistance (3), and regulatory networks in B-cell lymphoma (4), just to name a few.
The exchange of ideas is at the forefront of scientific advancement, and while publications serve as important libraries for documented research, the connections and discussions that take place at conferences are just as important. We were thrilled to participate in the American Association for Cancer Research (AACR) 2021 annual meeting and hear from researchers who are using single cell applications to transform the way we approach and understand cancer. Browse highlights from this event and additional related research at our content hub.
We invite you to keep the conversation going by joining us at the 10x Genomics Global Virtual Cancer Symposium, April 26–30. This weeklong webinar series offers the chance to continue learning from leading scientists as they share the progress they’re making with single cell technologies, including resolving the heterogeneity of the tumor microenvironment, exploring cancer progression, and developing a clearer picture of what drives successful treatment. Register today.
Feeling inspired for your own projects? What questions are you eager to ask, and how can single cells transcriptomics help you answer them? Where do you start when planning these experiments? Though there are many possibilities, don’t worry, we’re here to help—we’re excited to offer you this resource to help you get started and navigate key decision points along the way. Take a sneak peek before you download:
Getting started with Single Cell Gene Expression
Our planning guide walks you through the process of designing experiments that will best answer your research questions. To start, be clear about your goal. It may seem obvious, but knowing the specifics here will help determine the answers to all the workflow questions that follow.
How many cells do you need for your experiment? How many replicates? Some research questions can be sufficiently addressed with just one sample, while others may benefit from computational aggregates of different samples. Explore the variables, including sample diversity, the frequency of subpopulations of interest, and sample type, that impact the appropriate number of cells required for proper analysis and the potential need for biological replicates. Determine the sequencing depth you’ll need up front so you can get the answers you need the first time, without overspending on sequencing costs.
Generating sequencing data is only part of the equation. How do you turn this wealth of information into meaningful results? All single cell applications from 10x Genomics come with free, easy-to-use software for data analysis and visualization. Learn about Cell Ranger analysis pipelines that filter sequences, align reads, count barcodes, perform clustering and gene expression analysis, provide quality control metrics, and can now be run through 10x Genomics Cloud Analysis.* Output from Cell Ranger can be viewed interactively with Loupe Browser visualization software, which helps you identify cell types, rare subpopulations, and new marker genes, or imported to third-party tools.
Additional guidance can also be found in the form of use case examples. An organized table of references provides concise summaries of published work broken down by scientific aim, sample and library prep methods, sequencing metrics, and analysis tools.
Further exploration
We hope you find this resource to be helpful in your cancer studies and look forward to hearing about all the amazing advancements yet to come. We also encourage you to make the most of the ongoing conversations among oncology researchers, continuing to learn from the collaborative exchange of ideas at the upcoming Global Virtual Cancer Symposium, April 26–30. Register today.
*See 10x Genomics Cloud Terms of Use for restrictions and details
References
- Stewart CA, et al. Single-cell analyses reveal increased intratumoral heterogeneity after the onset of therapy resistance in small-cell lung cancer. Nat Cancer 1: 423–436, 2020.
- Wang R, et al. Single-cell dissection of intratumoral heterogeneity and lineage diversity in metastatic gastric adenocarcinoma. Nat Med 27: 141–151, 2021.
- Wu TD, et al. Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature 579: 274–278, 2020.
- Ott CJ, et al. Enhancer architecture and essential core regulatory circuitry of chronic lymphocytic leukemia. Cancer Cell 34: 982–995, 2018.
