Our 1.3 million single cell dataset is ready to download
At ASHG last year, we announced our 1.3 Million Brain Cell Dataset, which is, to date, the largest dataset published in the single cell RNA-sequencing (scRNA-seq) field. Using the Chromium™ Single Cell 3’ Solution (v2 Chemistry), we were able to sequence and profile 1,308,421 individual cells from embryonic mice brains. Read more in our application note Transcriptional Profiling of 1.3 Million Brain Cells with the Chromium™ Single Cell 3’ Solution.
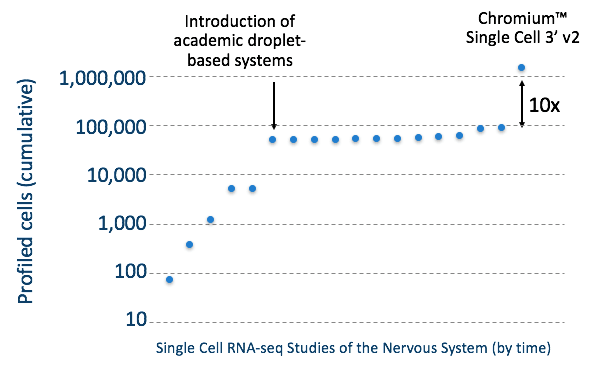
This dataset marks an order of magnitude increase, or, as we like to call it, a 10x increase, in the number of cell profiles generated via scRNA-seq than previously reported. While DropSeq brought about the first significant increase in the number of single cells profiled, the Chromium Single Cell 3’ Solution is responsible for the latest 10x increase, as illustrated in the graph below.
The Dataset
Cells from the cortex, hippocampus and ventricular zone of two embryonic mice were dissociated following the Demonstrated Protocol for Mouse Embryonic Neural Tissue and used to create 133 scRNA-Seq libraries. The samples were then sequenced on 11 Illumina Hiseq 4000 flow cells, resulting in a read-depth of approximately 18,500 reads per cell. The sequencing data were processed by Cell Ranger 1.2 to generate single cell expression profile of 1,308,421 cells as well as the clustering and differential gene expression analysis.
Watch the video of Tarjei Mikkelsen’s Million Cell Dataset announcement at ASHG 2016 to get a quick overview of the experiment.
Why it Matters
Cells are at the core of every living organism and, as such, understanding the fundamental components of life is key to understanding complex biological systems. The ability to profile a large number of cells is becoming increasingly important for rare cell detection, and for comprehensive classification of biological systems. However, existing scRNA-seq methods have limited throughput and lack scalable computational analysis tools. With the flexible throughput of the Chromium Single Cell 3’ Solution, the scalable Cell Ranger™ 1.3 analysis pipeline and Loupe™ Cell Browser visualization tool, it is now possible to start considering large-scale scRNA-seq experiments.
How will you use it?
That’s up to you! Our Million Cell Dataset defines a new standard for scaling up single cell analysis by orders of magnitude, opening up the possibility of tissue atlas studies that seek to comprehensively describe cellular subtypes and ultimately accelerate the characterization of all biological systems. To this end, we are making the dataset available for download without restrictions: Download the 1M cell dataset here.
Here are some tips about the dataset:
- You will find the gene-cell-barcode matrix consisting of all the brain cells, as well as the results of clustering analysis. Note that the "aggr – Gene/cell matrix HdF5 (filtered)" contains the filtered matrix in HDF5 format. Instructions to open the file in Python are here. We do not recommend loading the file into R, due to the file size and the lack of 64 bit integers support in R.
- We also provide a gene-cell-barcode matrix of randomly sampled 20k cells for a quick convenient browsing. This file can be opened in R using the function get_matrix_from_h5 in the Cell Ranger: R Kit.
- "Loupe Cell Browser file" can be used in the recently released Loupe Cell Browser to quickly and interactively identify significant genes, cell types, and substructure within the 1.3M brain cell data with no programming required. Learn more and dowload Loupe Cell Browser here.
- The raw FASTQ files are ~3.6 Terabytes in size, and will not be available directly from our website. Instead, we have uploaded them to Amazon S3 bucket from which users can download the files at their own cost using an Amazon account (depending on their region, Amazon may charge them up to $350 for this transfer). Interested users should follow the instructions here: /datasets. Bam files will be available from GEO soon.
