Rising to the challenge of complex biological questions: Greater insights into human health with high-throughput analysis
Our newest single cell instrument, Chromium X, increases throughput capabilities for a wide range of applications, enabling a new wave of biological insights. Find out how unlocking the power of scale can open doors to new understandings of disease pathology and accelerated therapeutic discovery, as we take a look at high-throughput data for exploring rare TCR clonotypes and antigen specificity in leukemia and the advancement of scalable, comprehensive drug screening techniques.
Strength in numbers
Think back to your early days in the lab. How many of the techniques that you use on a regular basis today hadn’t been developed yet? The rate at which methods in the biological sciences have evolved over time is impressive, yet this speed seems almost inevitable when we consider that these advances in technology are striving to keep pace with the ever-growing needs of the field—needs that arise as new research brings up more complex questions to answer. Single cell technology is one such advancement that has seen incredible growth, sustained over the past decade as researchers have discovered a wealth of information that could only be revealed by investigating biology at single cell resolution.
As the prevalence of single cell research expands to touch on all manner of fundamental questions relating to human health and disease, so too does the scope and scale of these studies. A 2020 survey of single cell transcriptomic publications shows a clear trend toward larger experiments (1). Along with a steady increase in publication rate is an increase in the number of cells being analyzed. The authors note that, “individual studies have increased in scale over time, and every few months, a new study is released that breaks the previous record in terms of number of cells assayed. During the first half of 2020, approximately 1,400,000 cells were added to the pool of public data every month.” A closer look at the results of these publications revealed that the number of cell types identified correlated strongly with the number of cells studied. Simply put, more cells means more data and an improved ability to refine cell subpopulations and detect more distinct cell clusters and rare cell types, all of which leads to a better understanding of the underlying biology of a sample.
Meaningful investigations of complex biological systems often require large single cell studies. Analysis of hundreds of thousands of cells enables patient cohort studies, deep profiling of a specific organ, and translational studies comparing healthy and diseased tissue. Scaling up to million-cell experiments allows for comprehensive cell atlassing, high-throughput drug and CRISPR screens, and deep immune repertoire analysis.
These demanding research objectives have long presented challenges. Reaching such ambitious cell numbers has been stymied by lack of streamlined workflows and the cost-prohibitive nature of investigating hundreds of thousands to millions of cells. Our latest single cell platform, Chromium X, provides the robust, scalable methods needed to not only make these types of studies accessible, but to make them routine. The second part of our Chromium X webinar series (available on demand) focused on high-throughput (HT) assays that can be run on the new instrument, with a deep dive into example datasets. Below, we’ll touch on highlights, including a look at TCR–antigen specificity and rare clonotype identification in leukemia and thorough characterization of drug response mechanisms in lung cancer cell lines.
Jump to: Drug screen data
Scaling up to tackle TCR repertoire diversity
Cancer is a formidable opponent, highly adaptable in nature and complex in its makeup. T-cell prolymphocytic leukemia (T-PLL) is a particularly aggressive form of cancer, characterized by out-of-control growth of mature T cells. Unfortunately, the vast majority of T-PLL patients do not reach long-term remission under standard treatments, making the discovery of novel therapies critically important for improving clinical outcomes. The incredible diversity of the TCR repertoire holds great potential for clinically relevant insights, and uncovering the details hidden within could provide direction for the development of new immunotherapy treatments (2). Using the Chromium Single Cell Immune Profiling HT assay with Feature Barcode technology, we examined TCR clonotypes and antigen specificity in a bone marrow aspirate from a T-PLL patient.
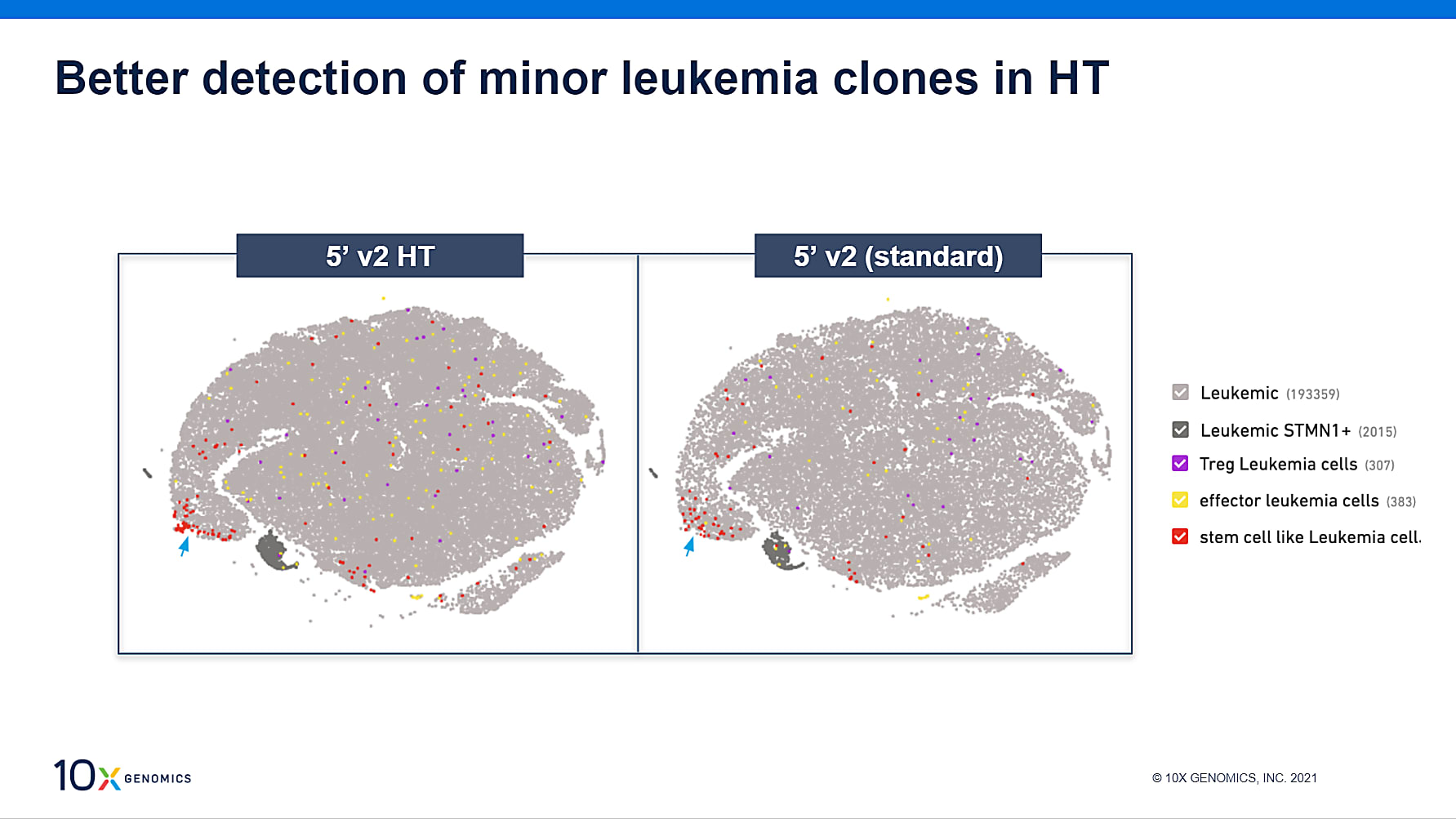
The HT assay was run in parallel with the standard assay, allowing for straightforward comparisons on the impact of throughput on the ability to identify rare cell types. While all leukemia subpopulations were identified with both kits, the higher cell throughput of the HT kit enabled better detection of minor clones, including the stem cell-like clone shown here in red.
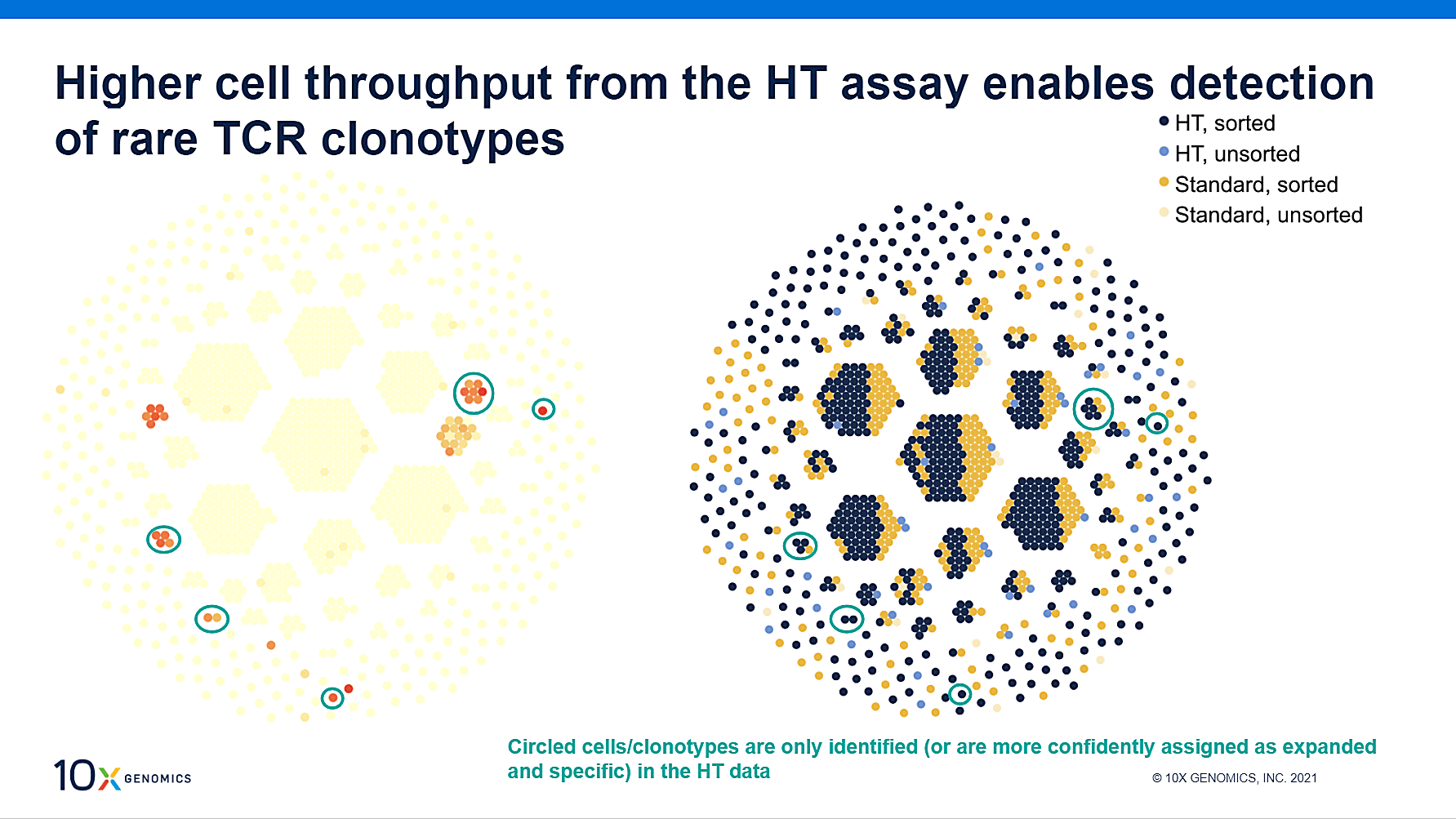
A panel of 27 dextramer reagents were used to stain the T-PLL sample, including control, viral, and cancer antigens. Only seven antigens were detected with a high median UMI count and evidence of antigen specificity. Of these, four were HLA-matched: two cancer antigens and two viral antigens. We took a closer look at one of the viral antigens, EBV, in order to assess confidence in antigen binding. If the same antigen binding is observed multiple times in multiple cells within a clonotype, confidence in binding assignment increases. As shown above, the HT assay provided data that allowed for the observation of more cells within the antigen-specific clonotypes, improving binding confidence and demonstrating better screening for rare antigen-specific TCR sequences.
Drug discovery from every angle with high-throughput screens
The drug discovery process is a laborious and time-consuming one, with disease complexity driving variable responses to drug treatment. Late-stage failures are common, making for a frustrating journey along the drug development pipeline. Even successful candidates take hundreds of millions of dollars and an average of seven years to reach the market (3, 4). High-throughput screening (HTS) enables the testing of a large library of candidate drug compounds, but in most cases, analysis is one-dimensional, offering single readouts such as cell morphology or activity of a single fluorescent gene at late time points. Unsurprisingly, this limited data provides minimal information about drug activity or mechanisms of action. Single cell HTS assays bring the opportunity to build a more complete picture of cell pathways impacted by the drugs in question. By collecting content-rich, single cell data on a massive scale, scientists can interrogate multiple treatments, time points, and/or cellular populations. This more comprehensive screen can better characterize the impact of therapies earlier in the drug pipeline.
We applied this principle to the examination of drug responses in two non-small cell lung cancer (NSCLC) cell lines, H1975 and A549. Optimal experimental conditions, including sample prep, drug concentrations, and treatment window, were set in a low-throughput (LT) experiment using the Chromium Single Cell Gene Expression LT kit. Each Chromium Single Cell assay utilizes the same chemistry and workflow, enabling a smooth transition from the pilot study to the full HT study. H1975 and A549 were each treated with four different drugs, some in combination, for a total of eight different treatments per cell line. Treated and untreated samples were collected at 4, 16, and 24 hours post-treatment. Cell multiplexing expanded screening capabilities beyond the capacity of even the HT kit alone, enabling analysis at appropriate scale for drug screening, with data collected on drug activity, effectiveness, and mode of action across 525,251 individual cells.

In H1975, two of the single drug treatments, crizotinib and osimertinib, effectively down-regulated cell cycle marker CDK2 at 16 hours post-treatment, and reached the same degree of downregulation as the combinatorial treatment with erlotinib after 24 hours. Crizotinib was chosen for further analysis into the biology driving this effect.
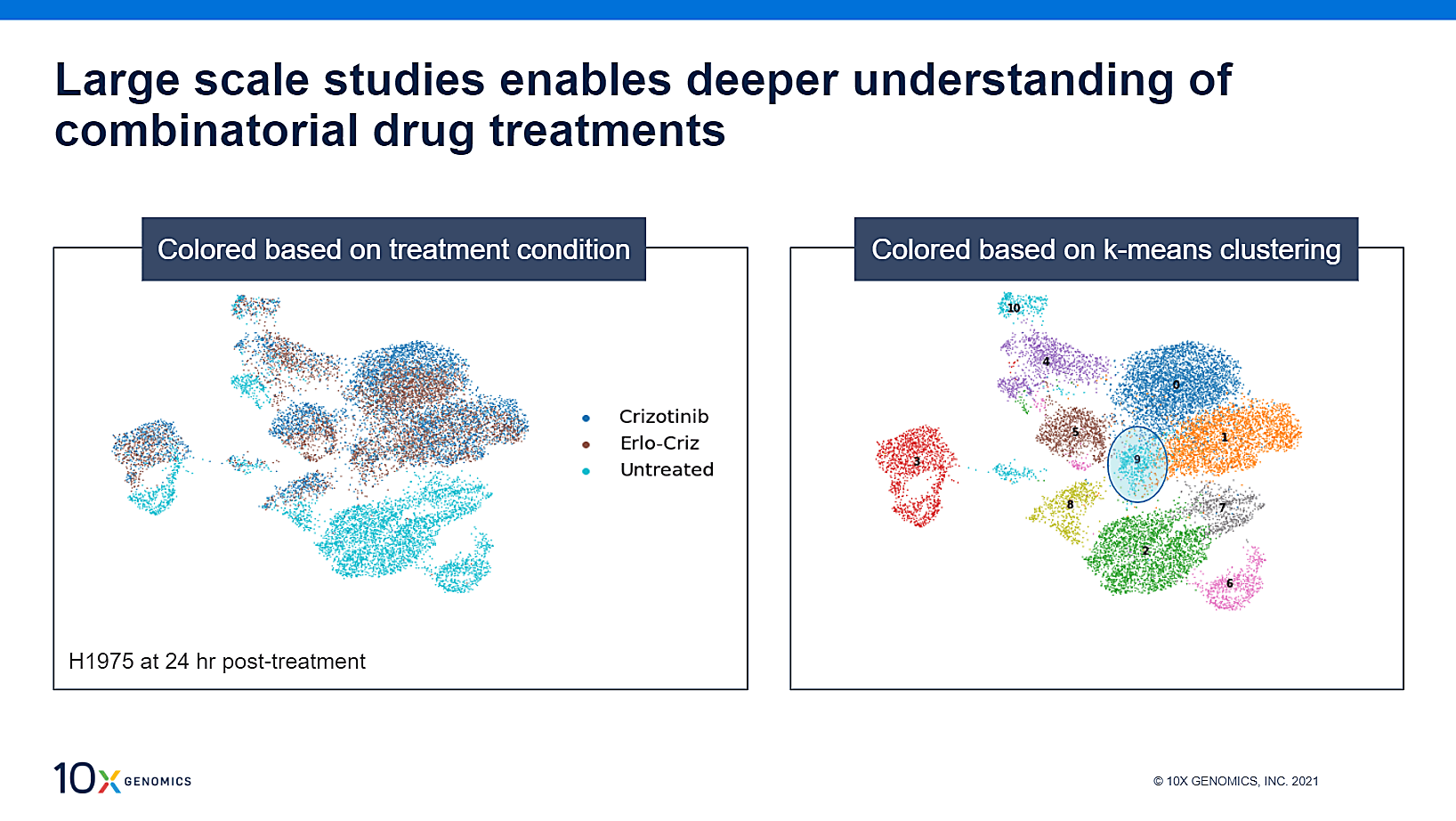
Further cell clustering analysis was performed for three treatment groups: an untreated control, crizotinib alone, and erlotinib in combination with crizotinib (erlo-criz). UMAP projection of the HT data (above) revealed 11 distinct clusters across the three conditions, with most crizotinib-treated cells overlapping with the erlo-criz treatment. Cluster 9 stood out from the rest, with twice as many erlo-criz-treated cells than crizotinib-treated cells, indicating a stronger response to the combinatorial treatment than was seen in the other clusters.
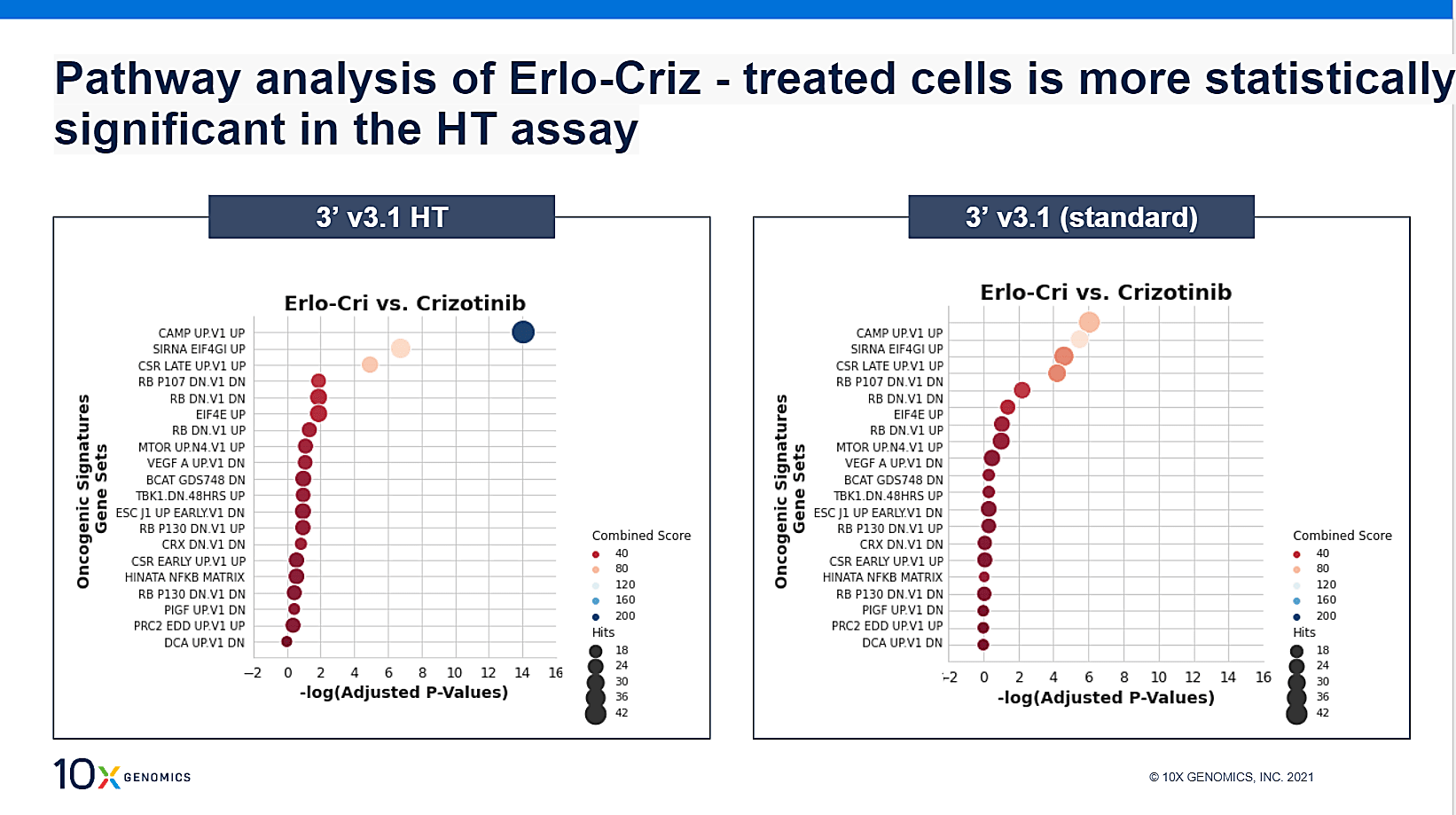
Closer examination of the gene expression profile of cluster 9 found that most up-regulated genes were mitochondria-related, pointing to cell stress. Gene set enrichment analysis (GSEA; above) showed the cAMP pathway, which regulates cell cycle, growth, and proliferation, to be significantly enriched. This suggested a mechanism for down-regulation of cell cycle genes, acting through the cAMP pathway. The same experiment was run in parallel using the standard Chromium Single Cell assay, and while pathway analysis was comparable, the higher cell throughput per sample of the HT assay boosted the statistical power of the analysis.
Let’s take a moment to return to the A549 cell line, carrying a KRAS mutation, used in the initial stages of this experiment. Notably, a separate earlier study looked at drug screen analysis of seven primary NSCLC patient samples, multiplexed together, most of which were known to also contain a KRAS mutation. The question naturally arose: how do the gene signatures compare between the A549 cell line and the patient samples?
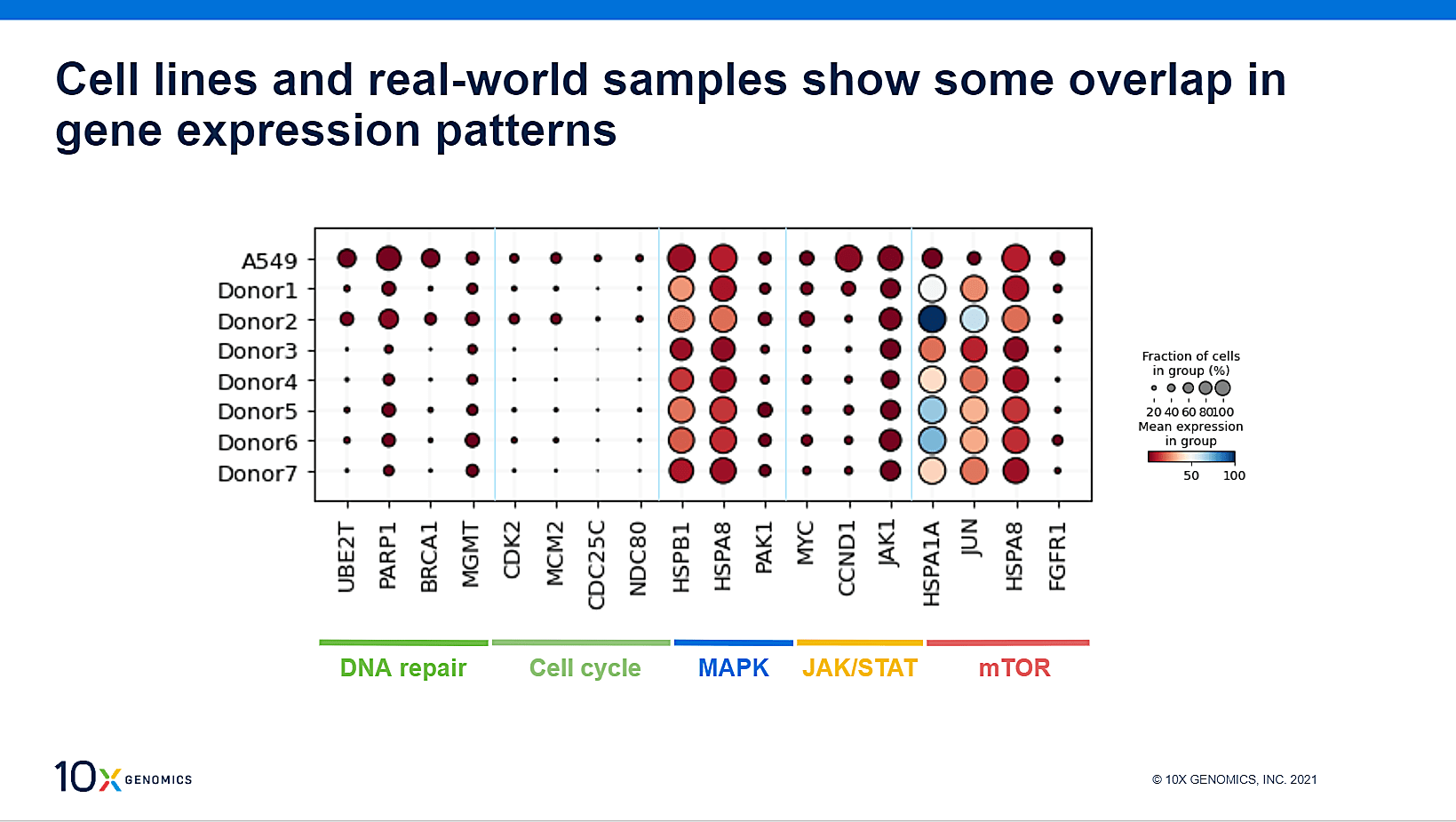
With both HT datasets at hand, we could detect distinct features and signaling overlays between the cell line and the real world samples, suggesting that A549 cells can be used for large-scale drug screens to identify new and more potent targets for KRAS-mutated NSCLCs.
**Jump to: TCR repertoire data **
Bringing new possibilities to the bench
We’re excited to see where HT applications can take research and how it can transform our understanding of human health and disease. These few examples are just the tip of the iceberg, and as new insights are revealed, more questions will inevitably follow. But if the recent past is any indication, technologies will advance to keep pace with increasingly ambitious needs.
That’s where Chromium X comes in, leveraging tried-and-true chemistry with increased throughput capacity to expand experimental possibilities and allow researchers to better answer critical questions about the mechanisms that underlie human health and disease. With these robust workflows, scientists can scale up their experiments in a cost-effective manner while maintaining the same sensitivity and multiomic outputs as our standard assays.
If you missed our Chromium X webinars, be sure to check out the series on demand to take a closer look at these datasets and hear more about instrument capabilities.
References:
- Svensson V, et al. A curated database reveals trends in single-cell transcriptomics. Database 2020, baaa073 (2020).
- Carter JA, et al. Single T Cell Sequencing Demonstrates the Functional Role of αβ TCR Pairing in Cell Lineage and Antigen Specificity. Front Immunol 10:1516 (2019).
- Prasad V, Mailankody S. Research and Development Spending to Bring a Single Cancer Drug to Market and Revenues After Approval. JAMA Intern Med 177(11):1569 (2017).
- Wouters OJ, McKee M, Luyten J. Estimated Research and Development Investment Needed to Bring a New Medicine to Market, 2009-2018. JAMA 323(9):844 (2020).
