FFPE Human Lung Cancer Data with Human Immuno-Oncology Profiling Panel and Custom Add-on
In Situ Gene Expression dataset analyzed using Xenium Onboard Analysis 2.0.0
Learn about Xenium analysis
Overview
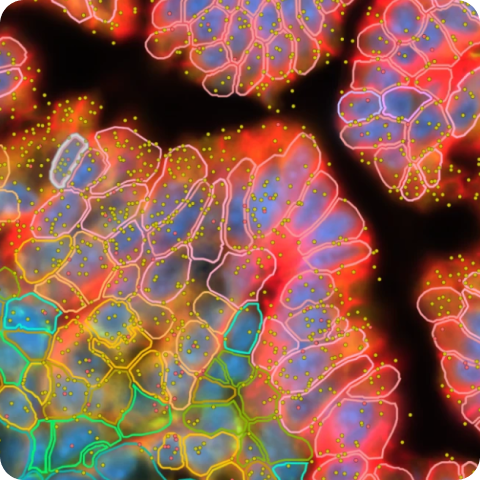
Xenium In Situ Gene Expression data for human lung cancer tissue using the Xenium Human Immuno-Oncology Profiling Panel with custom add-on.
How to view data
Interactively explore data with Xenium Explorer by downloading the Xenium Output Bundle (or Xenium Explorer subset) file. The subset bundle contains the experiment.xenium, gene_panel.json, morphology_focus/ directory of multi-file OME-TIFF files, analysis_summary.html, cells.zarr.zip, cell_feature_matrix.zarr.zip, transcripts.zarr.zip, and analysis.zarr.zip files.
See the Getting Started with Xenium Explorer page for more details. Follow these instructions to view the post-Xenium H&E image or image alignment file in Xenium Explorer.
Biomaterials
FFPE-preserved tissues were purchased from Discovery Life Sciences (Lung NSCLC; Stage I-B; Grade 2).
Tissue preparation
Tissues were prepared following the Xenium In Situ for FFPE - Tissue Preparation Handbook (CG000578). Probe hybridization, washing, ligation, and amplification were performed following the Xenium In Situ Gene Expression User Guide (CG000582).
Post-instrument processing followed the Demonstrated Protocol Xenium In Situ Gene Expression - Post-Xenium Analyzer H&E Staining (CG000613).
Gene panels
The Xenium Human Immuno-Oncology Profiling Panel (380 genes) was pre-designed by 10x Genomics. Genes were selected by a combination of data mining and manual curation (including literature review and domain expert feedback) and subsequent panel design was informed using single cell RNA sequencing data curated by the Tabula Sapiens cell atlas. Genes were chosen to allow identification of major immune cell types and subtypes, along with the inclusion of immune checkpoint molecules. This design enables delving deep into the cellular and subcellular levels of the tumor microenvironment, providing unparalleled insights into the behavior of both tumor and immune cells.
Xenium Analyzer
The instrument run was performed following the Xenium Analyzer User Guide (CG000584). The on-instrument analysis was run with Xenium Onboard Analysis version 2.0.0.
| Metric | Lung cancer |
|---|---|
| Median transcripts per cell | 131 |
| Cells detected | 161,000 |
| Nuclear transcripts per 100 µm² | 216.9 |
| High quality decoded transcripts detected | 32,069,615 |
| Region area (µm²) | 42,817,915 |
This dataset is licensed under the Creative Commons Attribution 4.0 International (CC BY 4.0) license. 10x citation guidelines available here.
