ASHG 2016 Round-up
Thanks to all of those who visited us at ASHG 2016 in beautiful (but not so sunny) Vancouver. For those of you who couldn't make it, here are the 10x highlights from the meeting.
HackSeq Hackathon
From Saturday to Monday 10x projects were worked on at the HackSeq Hackathon (with very little sleep, from what I hear). And the winner is…. 10x’s Grace Zheng and her team working on "Visualization of single cell RNA-seq data from tens of thousands of cells in real time". Learn more about the projects and HackSeq here: http://www.hackseq.com/.
New Chromium™ Single Cell Controller
We kicked off the week with an announcement about our new Chromium Single Cell Controller that brings affordable single cell analysis to any lab. Learn more about our new controller and Chromium™ Single Cell 3’ v2 Library and Gel Bead Kit, with >65% cell capture efficiency.
10x Community Mixer
Tuesday evening, we hosted a 10x Community Mixer to celebrate the launch of the online 10x Community. We would like to thank everyone who participated for helping make it a great success!

Wednesday, October 19: Agilent workshop and in-booth presentations
Wednesday afternoon started with the Agilent Workshop "Redefining the Exome: New Insights into Exons, Genes and Disease". David W. Craig from Keck School of Medicine of USC gave a talk about using 10x Genomics and Agilent’s OneSeq Exome Target Enrichment to generate phased exomes.

Following the workshop we hosted in-booth presentations with our own Sarah Garcia and Alex Wong. Sarah presented her ASHG reviewers choice poster "Linked-Reads for Improved Genomics", while Alex gave a great overview of Long Ranger Software for Linked-Read data analysis and then showed how to visualize the data during a Loupe Software demonstration. Datasets are available for download on the 10x Support Site.

Thursday, October 20: Posters, posters, posters, talks and more in-booth presentations
Two morning talks featured 10x technology. Click on talk title below to search for abstract by program number.
First up, Program No. 86 "High performance discovery of complex genome-wide rearrangements with single molecule-based barcoded sequence reads." Li Xia Stanford University
Second, Program No. 135 "Combining linked-read technology with standard target-enrichment NGS can accurately resolve short reads and distinguish variants in the Lynch/ CMMRD syndrome gene, PMS2, from its pseudogene, PMS2CL." Charlly Kao, Children’s Hospital of Philadelphia
The afternoon poster sessions highlighted 10x technology for exome sequencing and more. Click on the link to search for the absract by program number and day (Thursday 10/20).
- PgmNr 1133/T "High-coverage sequencing of the Human Genome Diversity Project (HGDP-CEPH) Panel." Shane McCarthy, Wellcome Trust Sanger Institute
- PgmNr 2537/T "Resolving the pseudogene mapping complexities of conventional exome sequencing in hearing impairment using 10x Genomics approach." Renata Pellegrino, Children’s Hospital of Philadelphia
- PgmNr 3185/T "Shining a light in the "dark matter" of the exome: Using linked-reads and exome bait capture to map NGS short reads to genes and their paralogous segments." Lifeng Tian, Children’s Hospital of Philadelphia
- PgmNr 3191/T ★ REVIEWERS CHOICE ★ "Long range applications with short read sequencing." Sarah Garcia, 10x Genomics
- PgmNr 3176/T "Advanced applications for clinical research exomes". Margherita Corioni, Agilent Technologies
- PgmNr 3188/T "Comparison of synthetic long read sequencing methods and optical mapping for de novo genome assembly." David Mohr, Johns Hopkins University School of Medicine
We ended the day with 2 in-booth presentations: "Single Cell RNA-Seq and Data Visualization" with Tarjei Mikkelsen and "Software Demo: Single Cells and Seurat" with Grace Zheng.

Friday October 21: 10x Workshop and 1 million single cell dataset!
The 10x Workshop "Transforming Genomic Analysis with the 10x Chromium System" featured speakers Dr. Chris Whelan, PhD, Broad Institute, Dr. Calvin Kuo, MD, PhD, Stanford University, and Dr. Tarjei Mikkelsen, PhD, 10x Genomics and was standing room only!

Chris Whelan discussed his work with the InPSYght project – producing WGS data for 10,000 schizophrenia, bipolar, and control samples to identify variants associated with disease, build a reference panel of structural events and haplotypes to impute into large genotype data sets, and create and share data and methods with the community. 10x Genomics Linked-Reads were able to not only detect additional structural variation, but also phase variants over long distances to determine haplotypes and also create reasonable de novo assemblies from a normal sample.
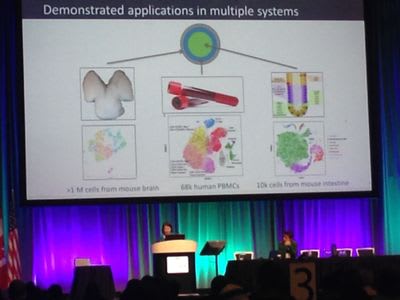
Tarjei Mikkelsen made some exciting announcements around our single cell applications including a million cell dataset and V(D)J paired sequencing.
After Tarjei, Calvin Kuo followed-up and spoke about his use of the 10x Genomics Single Cell Solution for studying the signaling pathways of intestinal stem cells (ISC). He focused in on Lrg5+ ISC and used single cell analysis to compare the gene expression profiles of control v. LOF and GOF mutants for two signalling pathways (Wnt and Rspo) implicated in Lrg5+ stem cell differentiation. He was able to conclude that the default state of Lrg5+ ISC is differentiation and self-renewal is an active process. The Wnt pathway confers a priming or competency step for Lrg5+ ISC renewal, while Rspo actively induces ISC renewal and specifies the size of the ISC pool. This research was able to demonstrate regulation of mammalian stem cells by by distinct priming and self renewal steps.
We’ll be doing more in-depth blog posts about our new 1.3 million cell dataset and V(D)J sequencing soon!
Recordings of the presentations will be available on the 10x website. We’ll update the blog post with the links once the videos are available.
Saturday, October 22: Wrapping up the meeting with 3 great talks
The party didn’t end on Friday, we still had 3 interesting talks on Saturday morning.
Click on the link to search for the abstract by program number and day (Saturday 10/22).
First up was Program No. 314 "A hybrid approach for de novo human genome sequence assembly, phasing, and detection of complex structural variation." with Yulia Mostovoy, Cardiovascular Research Institute, University of California, San Francisco.
Next 10x’s Grace Zheng spoke about Program No. 292 "Massively parallel digital transcriptional profiling of single cells".
Finally, we had Deanna Church’s talk "De novo assembly of individual human haplotypes from diploid samples."
Thanks for a great ASHG 2016! We’ll see you next year in Orlando at ASHG 2017.
