Earth Day 2018: A 10xer’s Take on Conservation Genomics
Guest Author: Andrew Gottscho, Software Field Operations Engineer, 10x Genomics
June 30, 2017. Green Cay National Wildlife Refuge, St Croix, U.S. Virgin Islands
The tropical sun beat down through the filtered canopy of stunted trees. During late morning at this latitude and season, the angle of solar radiation was nearly perpendicular. Through the undergrowth I could see patches of the cobalt Caribbean Sea glistening just a couple hundred meters away, hear the waves breaking upon the exposed reef at low tide, barely catching a breath of the salt-laced trade breeze. I wanted nothing more than to dive beneath the waves.
‘Maybe I should be preparing for my job interview with 10x Genomics instead of galavanting around this island - it’s less than two weeks away,’ my conscience berated me belatedly as I stood motionless in a scrap of shade, scanning the leaf litter for any sign of movement.
Yet I was not on vacation. I was a postdoctoral fellow here with my colleague Nicole Angeli from the Smithsonian Institution’s National Museum of Natural History on a field research trip to study the endangered St Croix Ground Lizard (Ameiva polops). Our goal was to collect tissue samples for population genomics analysis and translocate individuals to nearby Buck Island Reef National Monument, small (0.71 square km) and uninhabited, and one of the first protected coral reefs in the United States.

Under the supervision of U.S. Fish and Wildlife biologists and local authorities, we had paddled out on kayaks to Green Cay, a mere 0.06 square km, from the "main" island of St. Croix (236 square km). The Green Cay National Wildlife Refuge was established in 1977 to protect one of the last remaining populations of this endangered species. Ameiva polops was never widespread in the world, being endemic to St Croix, descended from Puerto Rican ancestors that presumably drifted here across the sea in prehistoric times. The introduction of the mongoose, a voracious mammalian predator widespread in the Old World, led to its extinction on St Croix. Now the species only exists on four tiny offshore islets where mongoose were never introduced.
In 2008, lizards from Green Cay were reintroduced by the U.S. Fish and Wildlife Service to nearby Buck Island. Because these tiny lizard populations might be susceptible to inbreeding depression, exacerbated by the risk of hurricanes, our aim was to mix up the gene pools by translocating individuals between islands. This "genetic rescue" technique was most famously used to save the endangered Florida Panther, stranded in the Everglades, from the effects of inbreeding depression when the U.S. Fish and Wildlife Service translocated just eight animals from Texas in the late 1990s (Johnson et al., 2010).
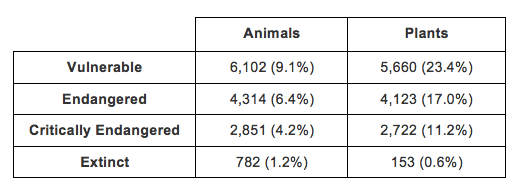
Sadly, the story of the St Croix Ground Lizard is common globally. Species are going extinct at alarming rates due to habitat destruction, invasive species, disease, and even climate change. Just a month ago, the last living male Northern White Rhino died at a Wildlife Conservancy in Kenya; with only two females of the species remaining, there is little hope for its survival (Gibbons, 2018). The IUCN Redlist has evaluated the conservation status of 67,222 species of animals and 24,230 species of plants (representing only a fraction of the unknown number of species on the planet):
Island natives, which frequently have nowhere else to go and often have lost their natural defences against continental predators, are especially vulnerable (for a review see Song of the Dodo: Island Biogeography in an Age of Extinction). The good news is that when scientists, government, industry, NGOs, and local communities work together, this pattern can be stopped or reversed - at least sometimes. For those like me who prefer seeing the glass half-full, see #EarthOptimism2018 on your social media platform of choice.
Famished after a week of successfully translocating dozens of lizards and collecting fresh tissue samples, we stopped by a local restaurant to snack on roast chicken and plantains. Trying to talk over the loud reggae music with our mouths full, Nicole asked me more about the genomics aspect of her project.
- How could we determine which genes were under directional selection, say due to predation by mongoose or climate change?
- How could we link alleles that show signals of selection to deleterious or advantageous phenotypes?
- How could we gauge the efficacy of our translocation efforts on the genetic health of the satellite populations to prevent inbreeding or disease?
- How could we control mongoose populations on the main island so that the lizards could eventually be reintroduced where they belong?
For the first three questions, I admitted that her existing restriction-associated DNA sequencing (RADseq) dataset that we collected, although valuable for many population genetics applications, was not enough to fully answer these questions. To link genotype and phenotype in a meaningful way, we needed a high quality, well-annotated reference genome that we could map her short RAD loci to. For the fourth question, we discussed gene drive, a new and controversial method of invasive species control on islands - something I knew almost nothing about, other than that it also required a good reference genome as a starting point.
Again, my thoughts turned to 10x Genomics. I knew that this small but rapidly growing biotech company was disrupting the genomics world by bringing down the cost of high-quality de novo assemblies using their patented Linked-Read technology, and more recently, single-cell genomics. They were bringing down the cost and technical barriers enough so that individual grad students or postdocs like us could potentially assemble a genome de novo, without a big NSF grant and a team of twenty collaborators!
My hope was to soon work for this emerging company that promised to not only personalize human medicine but to also provide valuable tools for biologists who study and manage non-model organisms - perhaps one day even the enigmatic St Croix Ground Lizard...
High Quality Reference Genomes Aid Conservation Efforts
You might be wondering, how exactly can high quality reference genomes and other biotechnology aid conservation efforts for endangered species? To start let’s briefly consider the genome of the African Cheetah (Acinonyx jubatus), a poster child for conservation and specifically for low genetic diversity among threatened species.
The world’s fastest land mammal, cheetahs evolved in North America and dispersed into Africa across the Bering Land Bridge during the "Ice Ages" of the Pleistocene epoch. This species is thought to have gone through at least two major population bottlenecks resulting in low genetic diversity. In modern times, its numbers have been further reduced by habitat loss and poaching, and it now only numbers a few thousand individuals in the wild. The cheetah as a whole species is listed as "Vulnerable" on the IUCN Redlist, but the subspecies A. j. hecki and A. j. venaticus are listed as "Critically Endangered."
Thus there has been great interest in breeding cheetahs in zoos, but this low genetic diversity comes with a price. Cheetahs are plagued by spermatozoa aberrations in males, elevated juvenile mortality, and increased vulnerability to infectious disease outbreaks.
Dobrynin et al. (2015) published the genome of the African Cheetah (in this case, a male named "Chewbacca") using short-read sequencing (75x coverage on a Illumina HiSeq2000) with seven mate-pair libraries. Assembled with SOAPdenovo2, they found a total genome size of 2.38 Gb, a scaffold N50 length of 3.1 Mb, and a contig N50 length of 28.2 kbp. They also resequenced several other cheetahs against their reference to get a sense of variation among individuals.
They found that the majority of the cheetah genome, as divided into 50 kbp windows, showed 8–15-fold less variation in cheetahs than in the human, domestic cat or wildcat. They also found that cheetahs had on average 10–15-fold longer homozygous stretches compared to the feral domestic cat genome (on average 93% of each cheetah’s genome was homozygous). Cheetahs lost 2–4 MHC class I genes, and near zero class I amino acid variation was found across seven cheetah genomes (unlike domestic cats which have appreciable MHC diversity).
Perhaps the most important finding was the discovery of 18 genes with damaging mutations implicated in spermatogenesis, azoospermia, oligospermia, gonadal dysfunction and oogenesis, and 10 fixed amino acid variants in the AKAP4 gene, expressed in the testis, which plays a critical role in sperm development and specifically of spermatozoa aberrations. These are almost certainly the "smoking guns" that have caused issues for zoo breeding efforts.
The key takeaway from the cheetah genome project is that it directly linked genotype to phenotype to fitness effects. These variants could, in theory, be targeted using gene editing (e.g. CRISPR-Cas9) to improve the success of captive breeding (Novak et al., 2018), just as gene editing is being used to design custom therapies for a variety of human diseases. In critical cases, this could mean the difference between extinction and survival. For example, the U.S. Fish and Wildlife Service is currently considering a proposal to use genetic engineering techniques to facilitate resistance to sylvatic plague in the black-footed ferret, which was presumed extinct but saved by a captive breeding program (Novak et al., 2018).
Harnessing the Power of CRISPR Technology to Help Endangered Species
Another application of CRISPR-Cas9 and genomics is using ‘gene drive’ technologies to override normal Mendelian inheritance, in which target deleterious alleles are inherited by >90% of offspring (Nolan 2018). Gene drive solutions have been proposed to fight mosquito-borne diseases, which affect not only humans but endangered wildlife as well, such as the endemic Hawaiian honeycreepers (Novak et al., 2018). We couldn’t help speculating about the possibility of replacing toxic chemicals, expensive fences and inhumane traps with more targeted, ecologically and economically friendly methods of controlling mongoose, rats, mice, and other invasive species that are wreaking havoc on island endemics around the planet.

Of course, to make use of CRISPR-Cas9 in a safe and controlled way for conservation purposes, a high-quality reference genome is a necessity. When the cheetah genome was published in 2015, short reads (Illumina) and long reads (PacBio, Oxford Nanopore) were the only practical options for genome assembly, and the bioinformatics was no breeze either. Short reads alone were not great, at least for assembling repetitive regions of the genome, or other difficult regions such as the MHC. Mate-pair libraries were better than short reads alone, but required additional technical expertise and expense. Long reads were pretty good - if you had lots of cash.
But if you wanted a high quality assembly, say with scaffold N50s measured in the megabases, at low cost, say only a few thousand dollars, you were still out of luck.
Enter 10x Genomics and Supernova, offering a complete de novo assembly solution with Linked-Reads.
January 13, 2018. Town and Country Hotel, San Diego, California
Just over six months after my job interview, I was now proudly representing 10x Genomics at the Plant and Animal Genome XXVI Conference, colloquially known as "PAG" amongst its loyal aficionados. Excitement was high as we were just about to launch Supernova 2.0, our de novo assembly software tool. Not only did our developers make numerous improvements in the algorithms, but we had successfully used Supernova to assemble twenty high quality human and non-human genomes, including a variety of plants, mammals, insects, and a bird. Although non-human assemblies are still considered to be experimental - please see Achieving Success with De Novo Assembly for precautions and caveats - I was excited to see our customers embracing our technology and boldly attempting to assemble genomes of organisms spanning the tree of life, from plants to animals and even metagenomes (something that is not supported at this time).
From the perspective of a conservation biologist, Supernova is attractive for the following reasons:
- Creating a genome assembly is easy. With the exception of DNA extraction, the assembly process is "pushbutton". There is only one parameter which you supply to Supernova: the number of reads to use, which can be computed from your estimate of the genome size.
- It only needs a tiny amount of DNA. This DNA needs to be high-molecular weight, but the low input requirement is important when working with endangered species where it may be difficult to obtain large volumes of tissue.
- It is cost-effective and scalable. Many conservation projects start off with small budgets, so the fact that a high-quality genome can be obtained for only a few thousand dollars cannot be overstated.
- It yields a high-quality, diploid representation of your genome. From the perspective of captive breeding and population genetics, it may be important to know phasing information.
Of course, Supernova 2.0 is just the next step in our journey. We have just launched our Single Cell CNV Solution and are planning to release new Feature Barcoding (including single cell CRISPR screens) and Single Cell ATAC-seq products. I have no doubt that our customers will delight and surprise us in how they apply these fundamental tools to solve pressing biological problems faced by society today.
Are you interested in conservation genomics or being a 10x customer? We’d love to hear more about your research interests and needs. Please email us at support@10xgenomics.com! And have a happy Earth Day!
References:
Gibbons, S., National Geographic Digital. 2018 March 20.
Johnson, W.E., et al., Science. 2010 Sep 24;329(5999):1641-5.
Dobrynin, P., et al., Genome Biol. 2015 Dec 10;16:277.
Novak, B.J., Maloney, T., and Phelan, R., The CRISPR Journal. 2018 Feb (1).
