Growing a Heart
Guest Author: Pia Abola
February 14 is traditionally a time for people around the globe to focus on their hearts, at least in a metaphorical sense. This year, it’s also a great time to reflect on the development of the heart in a biological sense, with the recent publication of a groundbreaking study that maps individual cells and their changing gene expression patterns during cardiogenesis (1).
The tools of modern molecular biology and biochemistry have given us amazing insights into developmental biology and have shown us how highly orchestrated patterns of gene expression are critical for embryogenesis and organogenesis. However, available technology has also limited our ability to visualize the full complexity of gene expression changes during this process. That’s why the recent study by Michaela Asp and colleagues at the KTH Royal Institute of Technology is so exciting. Combining advances in single cell gene expression profiling and spatial transcriptomics, the research team assembled a three-dimensional organ-wide atlas of the cells in the human heart at three different time points during development.
Insights Using Spatial Transcriptomics and Single Cell RNA-Sequencing
First, the authors collected four, nine, and six tissue sections along the dorsal-ventral axis of the heart from the 4.5-5, 6.5, and 9 weeks post-conception developmental time points, respectively. They explored global spatiotemporal gene expression patterns in the tissue using spatial transcriptomics, a technology that enables unbiased, high-throughput total mRNA analysis for intact tissue sections within the morphological context of the tissue. From this analysis, the authors identified 10 spatiotemporally conserved clusters of cells which were shown to map back to distinct anatomical regions within the three stages of the developing heart. For example, they observed a series of cell clusters shared across the three hearts corresponding to myocardial regions. These clusters increased in number with age, reflecting the heart’s growth developmentally.
The team then performed differential gene expression analysis between anatomical regions to better understand what genes characterize these areas of the heart and what functional role these areas play. They noted regional clusters enriched with genes involved in conduction, metabolism, oxygen transport, and endocrine activity, to name a few. They also noted certain regional clusters had enriched genes associated with the development of physical structures in the heart. For example, clusters 5, 6, and 7 were enriched in genes related to heart valve and septum development, offering insights into both where and through what biochemical pathways these structures may begin to form. Together, this analysis enabled a more rigorous molecular annotation of the anatomic regions of the developing heart, how they arise spatially and what functions they serve. And the team was able to determine that differences in gene expression seem to be more pronounced between regions of the heart, than between time points during development.
In an effort to understand these gene expression patterns at the level of single cells, the research team dissociated the cells from a new, biologically similar sample of 6.5 week post-conception tissue and performed single cell RNA-sequencing (scRNA-seq). Importantly, they had split the heart tissue in half between the atria and ventricles to distinguish cells from the upper and lower parts of the heart and analyze relative cellular diversity. Following this procedure, they performed dimensionality reduction and clustering of the gene expression profiles of the two single cell fractions and identified 15 cell clusters, which were further categorized into cell types based on known marker gene expression. This revealed many known cardiac cell types but also showed that certain clusters were only in the upper part of the heart, offering further insights into the process of development for the main part of the atria and other resident structures.
Creating a Spatial Cell Atlas of the Developing Human Heart
With both single cell and spatial gene expression data sets in hand, the research team aimed to resolve the cellular heterogeneity within the spatial clusters and assign spatial locations to the cell types identified with scRNA-seq. To do this, they utilized in situ sequencing (ISS) with a panel of 69 key marker genes identified from their spatial and scRNA-seq analysis or reported to be important for cardiac development. They re-clustered their existing scRNA-seq dataset with only the 69 gene panel, and performed ISS on replicate tissue sections for the three stages of heart development. Combining ISS techniques with psiSeq cell mapping algorithms (2), they created a probabilistic spatial cell map that confirmed the spatial distribution of scRNA-seq-defined cell types within the predicted spatial clusters.
Through ISS analysis, they were also able to clarify some of the spatiotemporal characteristics of and heterogeneity within the cell clusters identified by scRNA-seq. For example, they identified two cell-type subpopulations within cluster 14: cardiac neural crest cells expressing ISL1 and Schwann progenitor cells expressing ALDH1A1. Though both cells were present in the same region of the heart, analysis of these genes by ISS across the developmental time points revealed that cardiac neural crest cells were only present in the earlier stages, while Schwann progenitor cells were only seen in the later stages.
Additionally, the researchers were able to resolve the heterogeneity of three types of cardiomyocytes, also known as cardiac muscle cells. These cells compose the atria and ventricles of the heart and are involved in contractile function. While two types expressed marker genes for and localized to the atria and ventricles, respectively, the third type expressed MYOZ2 and FABP3, and localized to both the atria and ventricles. This cell type had not been previously described in humans, and its characterization points to the discovery power of integrated spatial and single cell gene expression data.
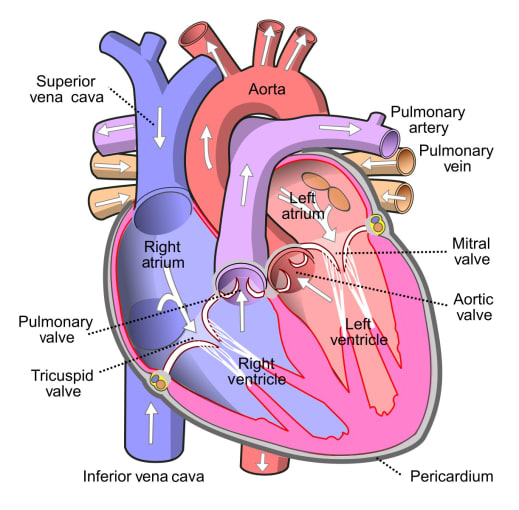
Biology is ultimately multidimensional and complex. Cardiac development demonstrates this fact, and points to the need for an analytical approach that can reveal spatiotemporal gene expression patterns within heterogeneous cell populations. Given the findings of this study, and the successful model of integrated spatial and single cell gene expression tools it offers, the road ahead for research on organ development is an exciting one.
The authors have provided publicly available web resources to explore the spatial gene expression patterns that regulate human cardiac development. You can access these tools here →
Other researchers are working to integrate spatial gene expression data with anatomical reference maps and single cell data. Watch this seminar to learn how Associate Professor Dinos Meletis and his research team created a molecular atlas of the adult mouse brain →
And if you want to learn more about cardiogenesis, read our blog post highlighting an important finding about the molecular mechanism underlying congenital heart defects. Read →
- M Asp et al. A Spatiotemporal Organ-Wide Gene Expression and Cell Atlas of the Developing Human Heart. Cell. 179 (7):1647–1660. (2019).
- X Qian et al. Probabilistic cell typing enables fine mapping of closely related cell types in situ. Nat. Methods. 17, 101–106. (2019).
