なぜ生物学に空間画像情報が必要なのか
シングルセル生物学は正常組織および異常組織の構成要素について特性解析を行う手段となることが明らかになっています。今日、細胞という構成要素から組織が形成される過程が空間生物学によって明らかにされたことから、組織に対する理解がさらに進みつつあります。これにより組織の地図帳を作り、希少な種類の細胞が存在する場所を特定することができます。さらに、細胞間の相互作用を視覚化し、細胞のシグナル伝達ネットワークを追跡し、バイオマーカーの特定が可能となります。そのいずれを行うときにも、空間的遺伝子発現の解析と空間プロテオミクスによって複雑な細胞組織系の探索が生物学的により意義深いものになります。
遺伝子発現の空間的特定、すなわち空間トランスクリプトミクスは、全トランスクリプトームの発現またはターゲットとする遺伝子の発現を組織切片内の固有の場所にマッピングした定量的な画像情報を生成する方法であり、本来の組織コンテキスト内における細胞の構成と活性を理解するために非常に有効であることが明らかになっています。
空間トランスクリプトミクスは、次世代シーケンスベースのアプローチで実現できます。このアプローチではmRNAを全トランスクリプトームレベルで組織にマッピングした後、ex vivoでmRANのシーケンスを行います。10x GenomicsのVisium 空間プラットフォームはこのアプローチを採用しています。空間トランスクリプトミクスは、数百から数千のmRNAをin situでイメージングする画像ベースのアプローチ(顕微鏡ベースの空間的方法とも呼ばれることもあります)でも実現できます。10x GenomicsのXenium In Situプラットフォームはこのアプローチを採用しています。空間的遺伝子発現画像情報を組織学的染色または免疫蛍光染色により同じ組織切片で検出されたタンパク質の情報と組み合わせると、組織の複雑さについてさらに深いインサイトが得られます。
空間トランスクリプトミクステクノロジーは、空間生物学、遺伝子プレックスレベル(検出された遺伝子の数)、遺伝子の感度(正確な転写物検出の線形ダイナミックレンジ)などの重要な基準によって評価されます。10x Genomicsの空間ツールは最高レベルのプレックスと感度で組織の複雑さに対する頑健なインサイトが得られるように最適化されているため、実験結果の信頼度が高まり、あらゆるサンプルに対してより深い理解が得られます。
空間トランスクリプトミクスのパワー
空間トランスクリプトミクスで何が可能なのでしょうか。例えば、以下に挙げる高インパクトの論文で示されているように、シングルセル研究プロジェクトに厚みを加えることができます。
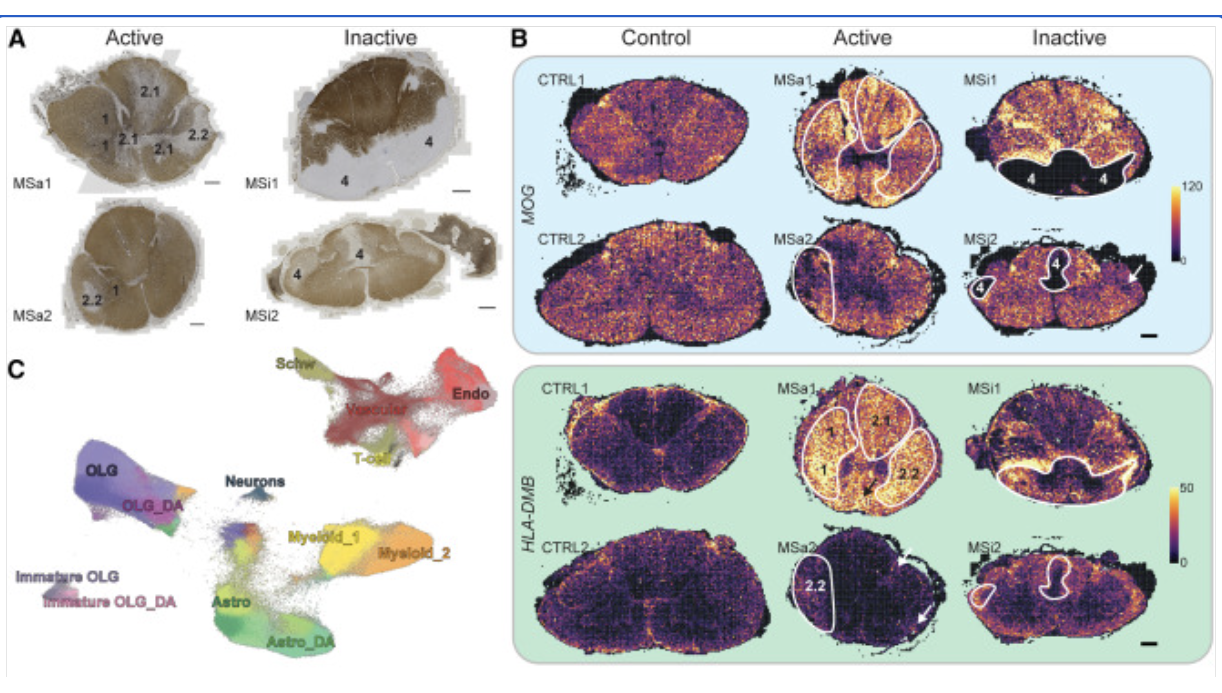
Xenium In Situを用いた多発性硬化症(MS)のヒト死後脳・頸髄、マウス死後脳の研究
Kukanja P, et al. Cellular architecture of evolving neuroinflammatory lesions and multiple sclerosis pathology. Cell 187: 1990–2009.e19 (2024). doi: 10.1016/j.cell.2024.02.030
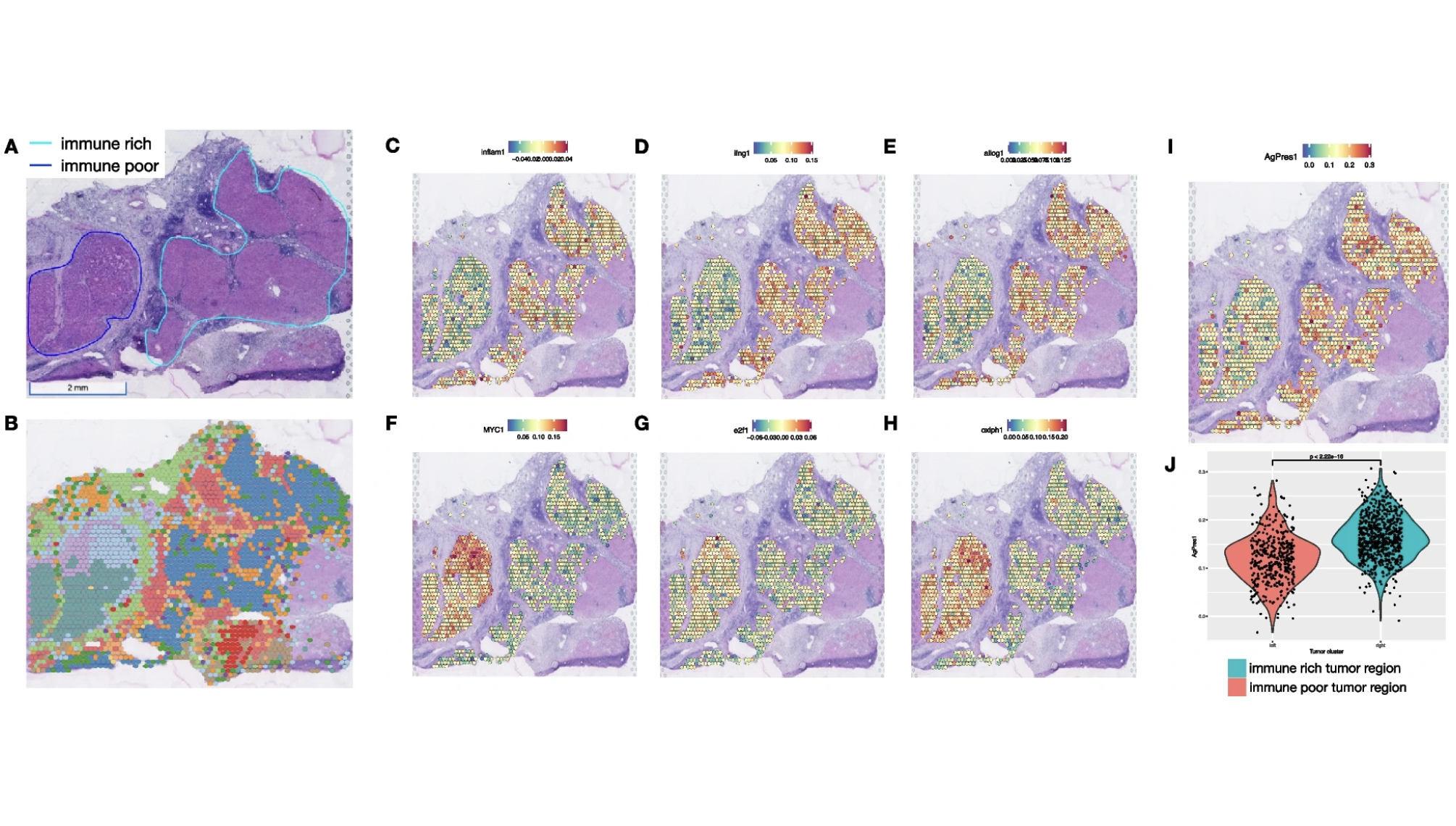
Visium Spatialを用いたヒト肝がんの研究
Zhang S, et al. Spatial transcriptomics analysis of neoadjuvant cabozantinib and nivolumab in advanced hepatocellular carcinoma identifies independent mechanisms of resistance and recurrence. Genome Medicine 15: 72 (2023). doi: 10.1186/s13073-023-01218-y

Xenium In SituとVisium CytAssistを用いたヒト胎児肺の研究
Quach H, et al. Early human fetal lung atlas reveals the temporal dynamics of epithelial cell plasticity. Nat Commun 15: 5898 (2024). doi: 10.1038/s41467-024-50281-5
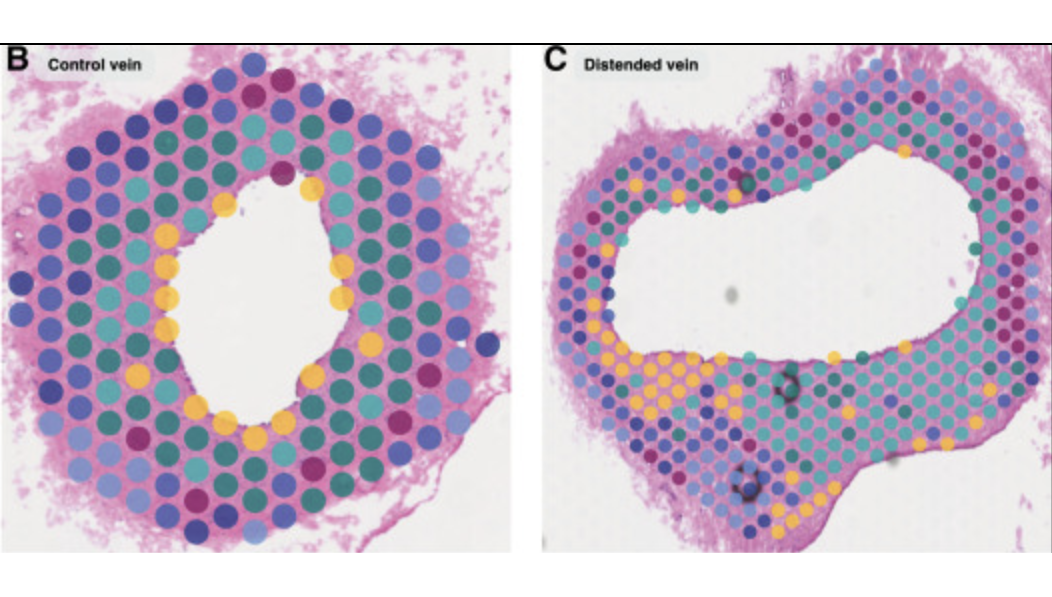
Visium Spatialを用いたイヌの心臓および静脈の研究
Michaud M, et al. Early injury landscape in vein harvest by single-cell and spatial transcriptomics. Circ Res 135: 110–134 (2024). doi: 10.1161/CIRCRESAHA.123.323939
2つのプラットフォームを合わせたパワフルな空間生物学ツールキット
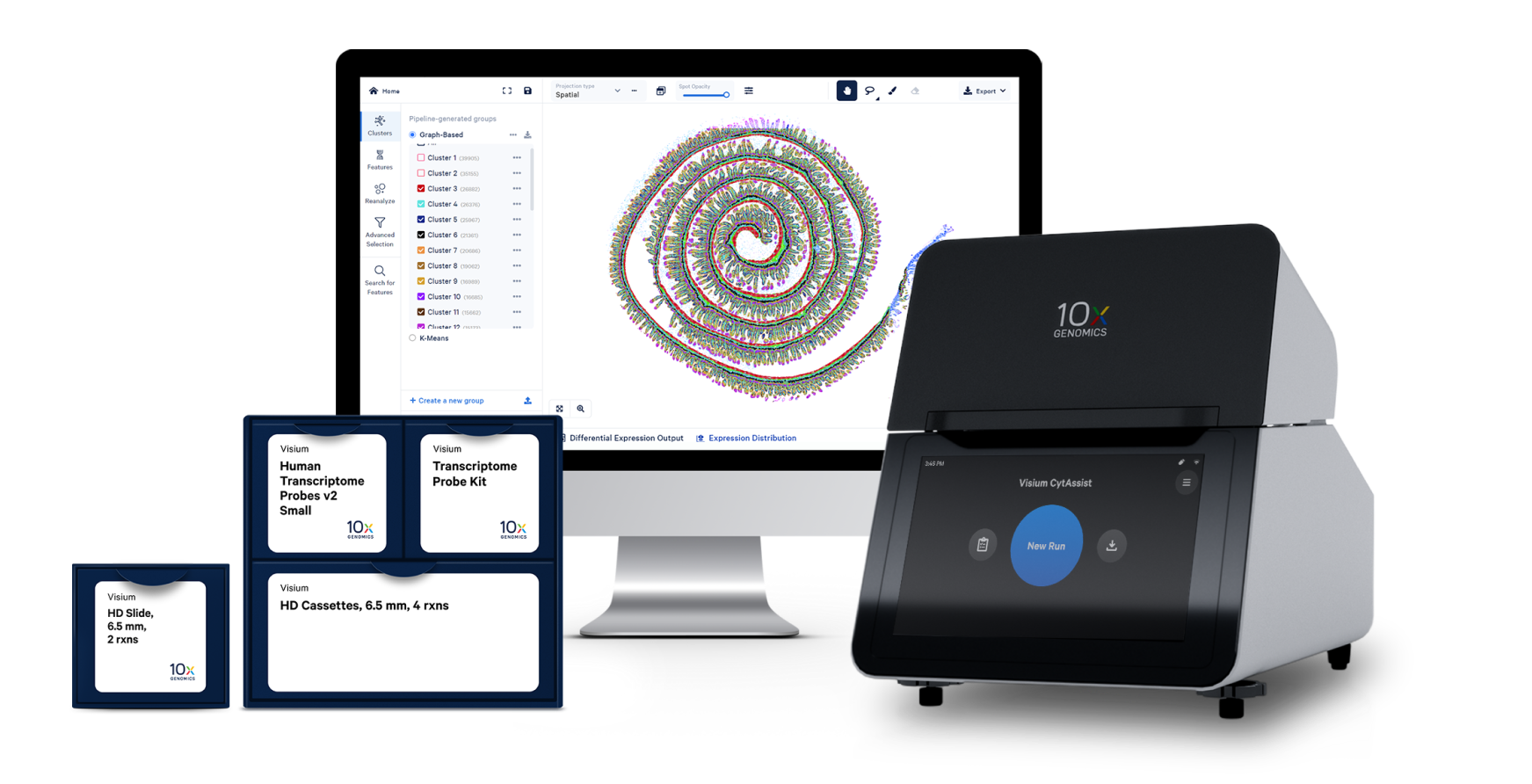
高解像度で全トランスクリプトームを空間的に特定します。
超精密シングルセル空間イメージングにより、最大5,000の遺伝子の特性を解析します。
空間データの解析
Visium空間解析とXenium In Situの両方でデータ探索を直観的に行えるため、空間データの解析時間が短縮されます。
シーケンスの生データを、明視野および蛍光顕微鏡画像と整合させた遺伝子発現データに変換します。
数回クリックするだけで、数千の空間的に可変な遺伝子を視覚化して調べられます。
組織コンテキスト内の遺伝子発現クラスターを調べ、細胞型や遺伝子シグネチャーを領域および形態により同定します。
オンボード解析と並行してXeniumを使用することでデータ探索の時間が短縮されます。
バイオインフォマティクスの経験がなくても、組織切片から得られたデータの視覚化プロセス全体を直観的に行えます。
組織中の特定の転写物について細胞および核境界内の位置を単一細胞解像度で正確に特定します。
実績
ビデオ
論文
FAQ
Vsium 空間解析は、どのアッセイを使用するかにより、さまざまな組織や動物種に対応します。新鮮凍結組織に対応するVisium空間遺伝子発現解析はポリ(A)ベースのキャプチャケミストリーを使用していることから、動物種にとらわれません。CytAssistを使ったVisium空間遺伝子発現解析は、ヒトまたはマウスサンプルの新鮮凍結、既固定凍結、ホルマリン固定パラフィン包埋(FFPE)組織切片について解析を行えるプローブベースのアッセイです。Visium HDが現在対応しているのはヒトまたはマウスのFFPE組織ですが、今後は対応サンプルの範囲を他の組織条件にも拡張することを予定しています。
Xenium In Situは現在、ヒトまたはマウスの新鮮凍結またはFFPE組織に対応しています。他の動物種についてはフルカスタムパネルをご利用いただけます。
10x Genomicsのサポートサイトで、Visium HD組織調製法(および他のVisiumアッセイへのリンク)やXenium組織調製法に関する関連文書もご覧いただけます。
VisiumアッセイもXeniumアッセイも、さまざまな種類の組織について検証済みの汎用サンプル調製ワークフローで最適化されています。しかし、固定し包埋するサンプルの調製プロセスは組織によって異なることがあります。Visium 空間解析とXenium In Situの両方がインプットサンプルとしてFFPEサンプルに対応しています。通常、FFPEサンプルを調製する際には、組織の構造を維持しRNA分解を制限するために、組織の切り出しから固定までの時間を最小限に抑えることをお勧めします。新鮮サンプルは慎重に扱うこと、固定前に最適な保存条件を整えることに特に注意してください。また、VisiumまたはXeniumでFFPE組織を用いた実験を開始する前に、FFPEブロックのRNA品質を検査することをお勧めします。詳細と推奨事項については、10x Genomicsのナレッジベースの記事をご覧ください。.
VisiumもXeniumもH&E染色に対応しており、画像ベースの組織形態画像情報が得られます。免疫蛍光染色にも対応しており、タンパク質発現画像情報を一定のプレックスレベルまで得られます。
また、Xenium In Situ マルチモダール細胞セグメンテーションは、自動細胞セグメンテーションへのインプットサンプルを染色するマルチ組織染色ミックスで、細胞膜や細胞内部をラベリングする抗体染色のほか、DAPI核染色も行えます。この染色ワークフローを用いてXeniumのランを実行すると、トランスクリプトームデータの形態マッピングをより正確に行えます。
VisiumでもXeniumでも同じ組織切片でタンパク質解析を行えるため、空間マルチオミックスを実現できます。ただし、プラットフォームごとにプロセスが異なります。
Visium HDを含むすべてのVisiumアッセイに、ランの前に組織切片のH&E染色または免疫蛍光(IF)染色を行うオプションがあります。したがって、IF染色でタンパク質検出を同時に行え、下流で空間的遺伝子発現データと重ね合わせた蛍光顕微鏡画像を得ることができます。さらに、CytAssistを使ったVisium空間遺伝子発現とタンパク質解析キットを使用するオプションがあり、キュレーション済み、検証済みの抗体コンテンツパネルを使用して単一の組織切片でタンパク質とRNAの両方を検出できます。
Xeniumでは、ワークフローの最後でH&E染色とIF染色を行えます。
VisiumはSpace RangerとLoupe Browseという2種類のソフトウェアでデータの解析をサポートします。Space Rangerは空間的遺伝子発現情報を組織画像と自動的に重ね合わせ、類似した転写プロファイルを示すバーコードのクラスターを同定する解析ソフトウェアです。次に視覚化ソフトウェアLoupe Browserが結果をインタラクティブに探索します。10x GenomicsのサポートサイトでSpace RangerとLoupe Browserを無料でダウンロードできます。
Xenium Analyzeはラン中にデータを自動的に処理するオンボード解析を行うことから、ラン後処理のために何時間も待つ必要はありません。その後、直観的な視覚化ソフトウェアXenium Explorerがin situデータをナビゲートします。
また、10x Genomicsの空間データアウトプットは、継続的に開発が行われている他社のツールと互換性を持つように設計されています。