Why biology needs a spatial readout
Single cell biology has proven to be instrumental in characterizing the building blocks of normal and diseased tissues. Now, spatial biology is further advancing our understanding of tissues by revealing the organization of those cellular building blocks. Whether atlasing tissues, localizing rare cell types, visualizing cell–cell interactions, tracing cell signaling networks, or revealing biomarkers, see how spatial gene expression and spatial proteomics can confer deeper biological meaning to your exploration of complex cellular tissue systems.
Spatially resolved gene expression, or spatial transcriptomics, is a quantitative readout of either whole transcriptome or targeted gene expression mapped to specific locations in a tissue section, and a proven powerful method to understand cellular composition and activity in the native tissue context.
Spatial transcriptomics can be achieved through next-generation sequencing-based approaches, where mRNA is mapped in tissues at the whole transcriptome level and then sequenced ex vivo. Our Visium Spatial platform leverages this approach. Spatial transcriptomics can also be achieved through imaging-based approaches—often referred to as microscopy-based spatial methods—where hundreds to thousands of mRNAs are imaged in situ. Our Xenium In Situ platform leverages this approach. A spatial gene expression readout can be combined with a histological stain or immunofluorescence protein detection in the same tissue section to elevate insights into tissue complexity.
Spatial transcriptomics technologies are assessed by key metrics, such as resolution of the spatial biology, gene plexity (the number of genes detected), and gene sensitivity (the linear dynamic range of accurate transcript detection). 10x Genomics spatial tools are optimized to provide robust insights into tissue complexity with utmost plexity and sensitivity, providing confidence in your results and a deeper understanding of every sample.
The power of spatial transcriptomics
Discover what's possible with spatial transcriptomics, including the additional depth it can add to single cell research projects, through these high-impact publications.
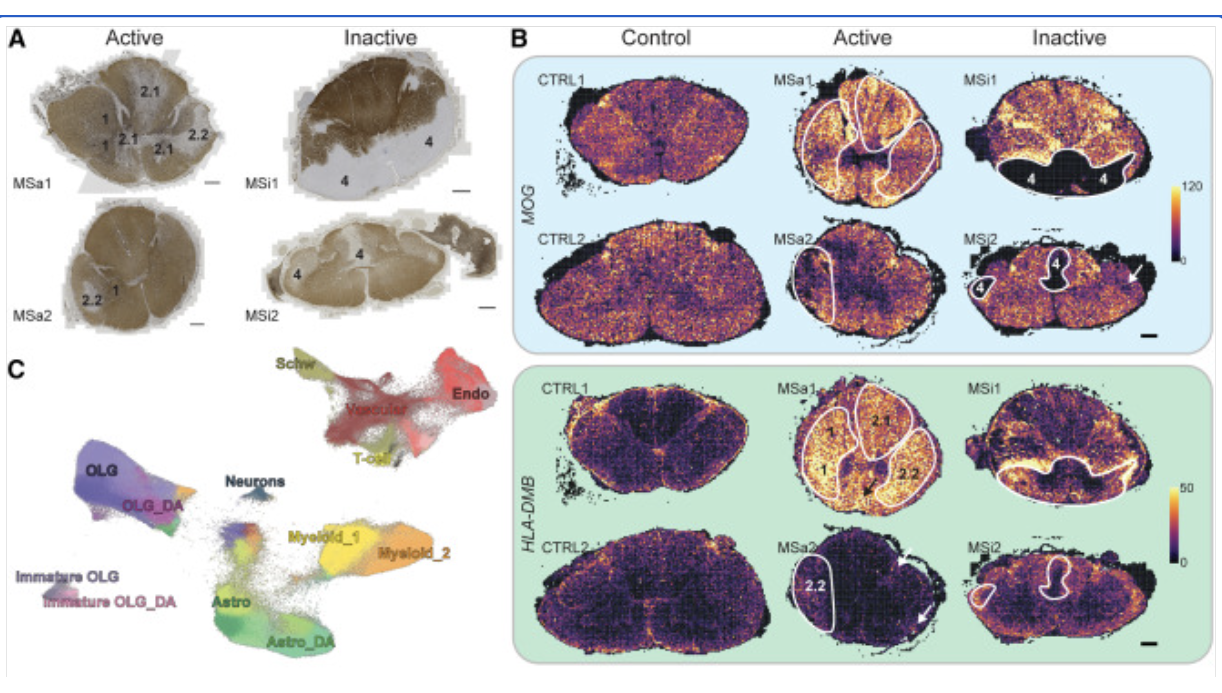
Human postmortem MS brain, cervical spinal cord; Mouse brain; Xenium In Situ
Kukanja P, et al. Cellular architecture of evolving neuroinflammatory lesions and multiple sclerosis pathology. Cell 187: 1990–2009.e19 (2024). doi: 10.1016/j.cell.2024.02.030
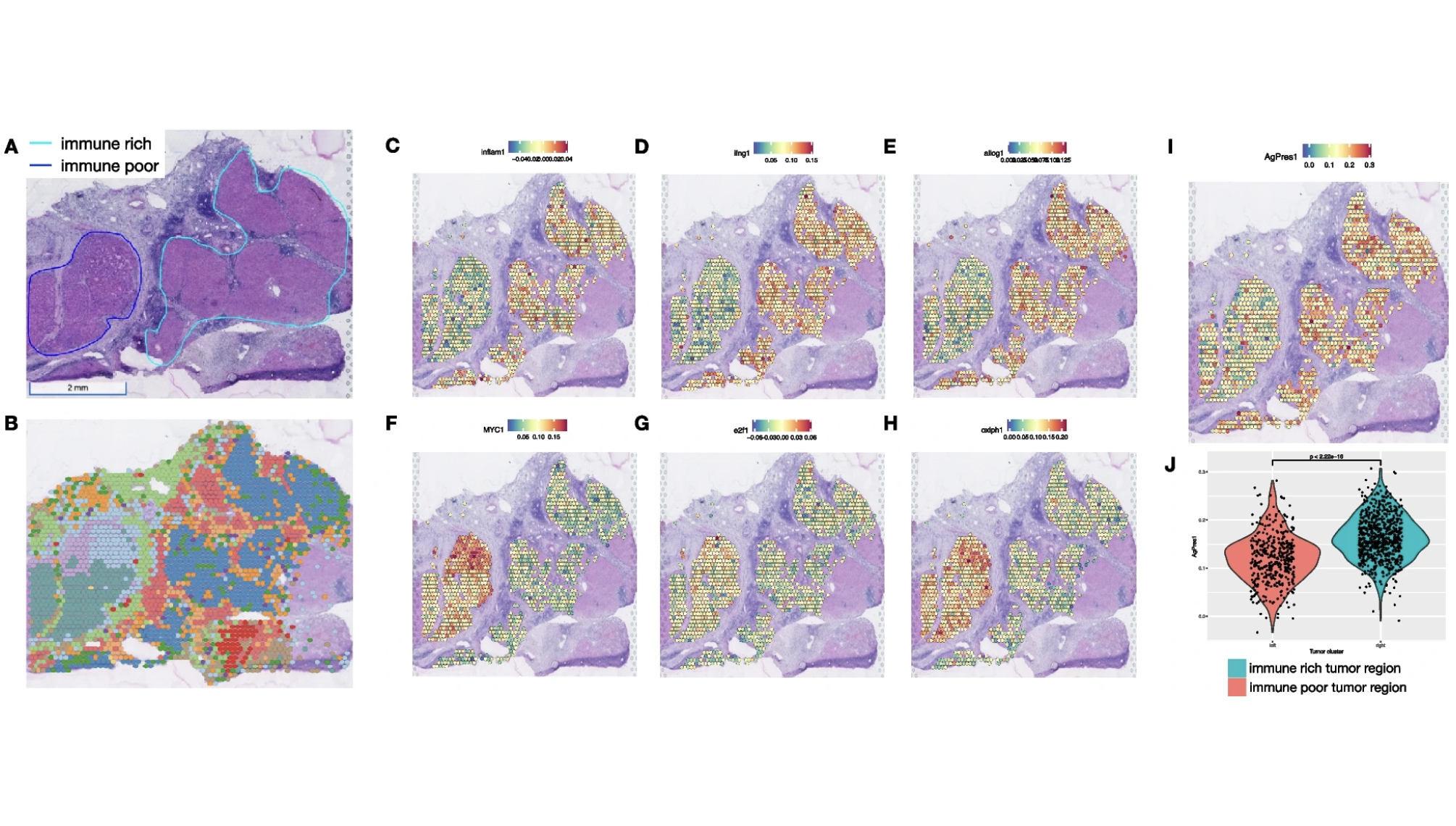
Human liver cancer; Visium Spatial
Zhang S, et al. Spatial transcriptomics analysis of neoadjuvant cabozantinib and nivolumab in advanced hepatocellular carcinoma identifies independent mechanisms of resistance and recurrence. Genome Medicine 15: 72 (2023). doi: 10.1186/s13073-023-01218-y

Human fetal lung; Xenium In Situ, Visium CytAssist
Quach H, et al. Early human fetal lung atlas reveals the temporal dynamics of epithelial cell plasticity. Nat Commun 15: 5898 (2024). doi: 10.1038/s41467-024-50281-5
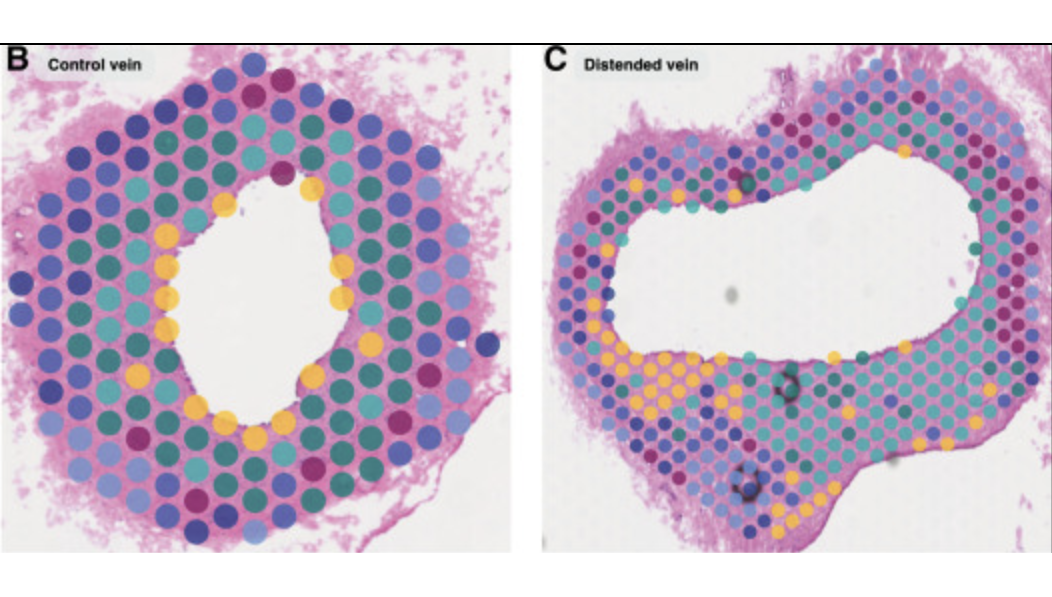
Canine heart, vein; Visium Spatial
Michaud M, et al. Early injury landscape in vein harvest by single-cell and spatial transcriptomics. Circ Res 135: 110–134 (2024). doi: 10.1161/CIRCRESAHA.123.323939
Two platforms, one powerful spatial biology toolkit
Whole transcriptome spatial discovery at high definition.
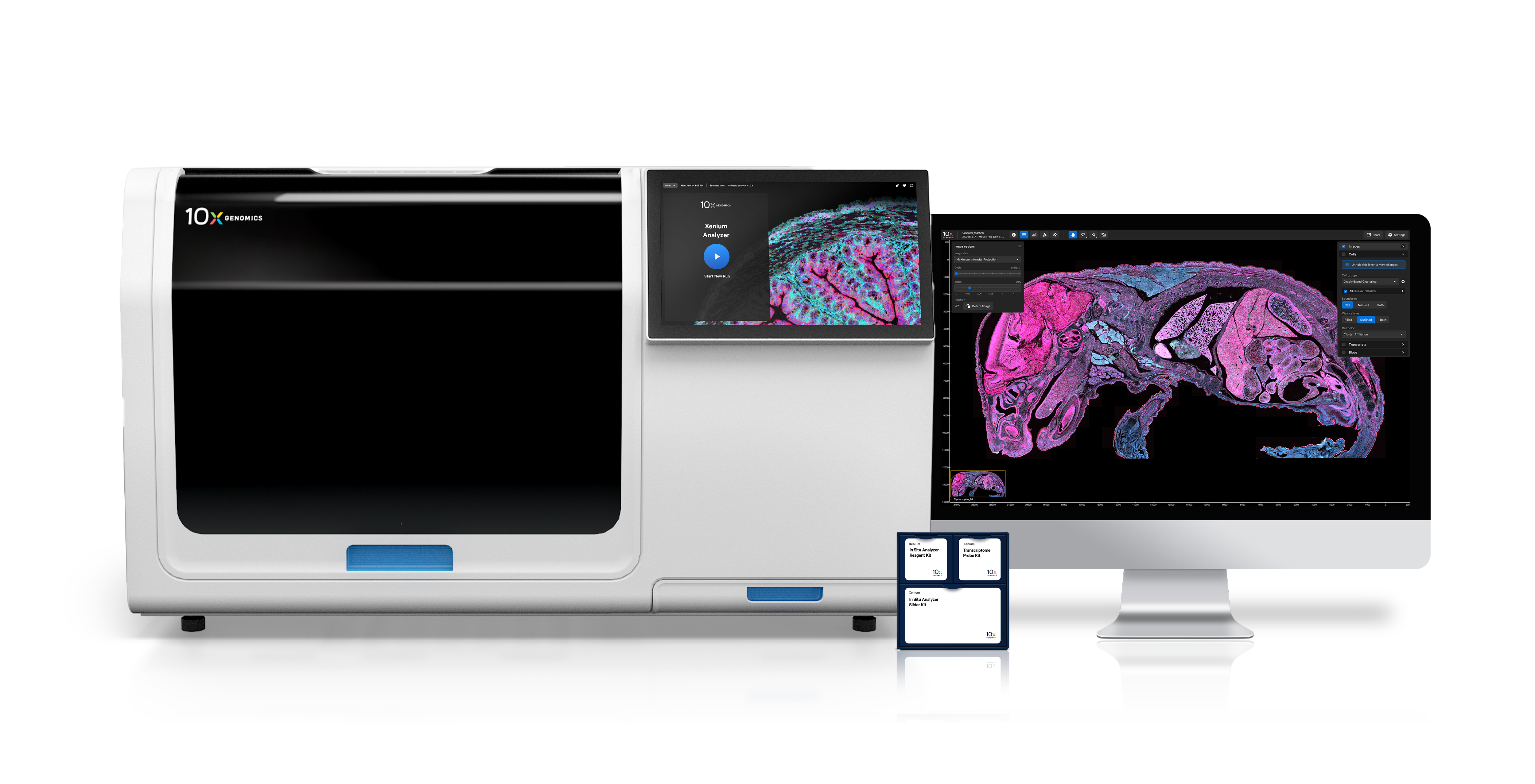
Characterize up to 5,000 genes with ultra-precise single cell spatial imaging.
Spatial data analysis
Accelerate spatial discoveries with intuitive data exploration for both Visium Spatial Gene Expression and Xenium In Situ.
Transform raw sequencing data into gene expression data aligned with brightfield and fluorescence microscope images.
Visualize and explore thousands of spatially variable genes in a few clicks.
Explore gene expression clusters in the tissue context to define cell types or gene signatures by region and morphology.
Run onboard analysis in parallel with the Xenium run to streamline time to data exploration.
Intuitively visualize data from each tissue section in its entirety, no bioinformatics experience required.
Pinpoint the locations of specific transcripts in tissues at subcellular resolution within outlines for cells and nuclei.
Proven results
Videos
Publications
Frequently Asked Questions
Visium Spatial is compatible with a variety of tissues and species, depending on the specific assay you choose to employ. Visium Spatial Gene Expression for fresh frozen tissue leverages a poly(A)-based capture chemistry and is therefore species agnostic. Visium CytAssist Spatial Gene Expression is a probe-based assay that enables analysis of fresh frozen, fixed frozen, and formalin-fixed paraffin-embedded (FFPE) tissue sections from human or mouse samples. Finally, Visium HD is currently compatible with human or mouse FFPE tissues, though we plan to expand sample access to other tissue conditions in the future.
Xenium In Situ is currently compatible with human or mouse fresh frozen or FFPE tissue sections. You can also build a fully customized panel for use with different species.
You can find additional resources for Visium HD tissue preparation (and links to other Visium assays) and Xenium tissue preparation on our support site.
Both the Visium and Xenium assays are optimized with a universal sample preparation workflow that is verified on multiple tissue types. However, preparing samples for fixation and embedding can be a variable process depending on the tissue. Generally, when preparing FFPE samples, which are a supported input sample type for both Visium Spatial and Xenium In Situ, we recommend minimizing the time between tissue resection and fixation to maintain tissue structure and limit RNA degradation. Special attention should be given to handling fresh samples with care and to optimizing storage conditions prior to fixation. Moreover, we recommend testing the RNA quality of FFPE blocks before starting either a Visium or Xenium experiment with that sample. You can find more detailed recommendations in our knowledge-base articles.
Yes, both Visium and Xenium are compatible with H&E staining, which provides an image-based readout of tissue morphology, and immunofluorescence staining for a limited plex protein expression readout.
Additionally, Xenium In Situ Multimodal Cell Segmentation is a multi-tissue stain mix that provides input for automated cell segmentation, including antibody staining to label the cell membrane and cell interior, as well as a DAPI nuclear label. This staining workflow supports enhanced accuracy in the morphological mapping of transcriptomic data from a Xenium run.
Yes, for both Visium and Xenium you can perform protein analysis on the same tissue section to achieve spatial multiomics. However, the process differs between the platforms.
For all of our Visium assays, including Visium HD, you have the option to perform H&E or immunofluorescence (IF) staining on your tissue sections prior to running the assay. IF staining thus enables simultaneous protein detection, and can be visualized downstream as a fluorescence microscope image overlaid with spatial gene expression data. Additionally, you have the option of utilizing the Visium CytAssist Spatial Gene and Protein Expression kit to study both protein and RNA from a single tissue section using a curated, pre-validated antibody content panel.
For Xenium, H&E and IF staining can be performed at the end of the Xenium workflow.
Visium provides two types of software to help you analyze your data: Space Ranger and Loupe Browser. Space Ranger is an analysis software that automatically overlays spatial gene expression information on your tissue image and identifies clusters of barcoded squares with similar transcription profiles. You can then use Loupe Browser, a visualization software, to interactively explore the results. You can download Space Ranger and Loupe Browser from the 10x Genomics Support site at no cost.
Learn more about Visium Spatial analysis
The Xenium Analyzer provides onboard analysis that automatically processes data during a run, without needing to wait for hours of post-run processing. Subsequently, you can use Xenium Explorer, our intuitive visualization software, to navigate your in situ data.
Learn more about Xenium In Situ analysis
Moreover, 10x Genomics spatial data outputs are designed for compatibility with third-party tools, which are in continuous development.