Whole transcriptome spatial discovery
Best-in-class spatial data quality with complete coverage

Visium slides and reagents
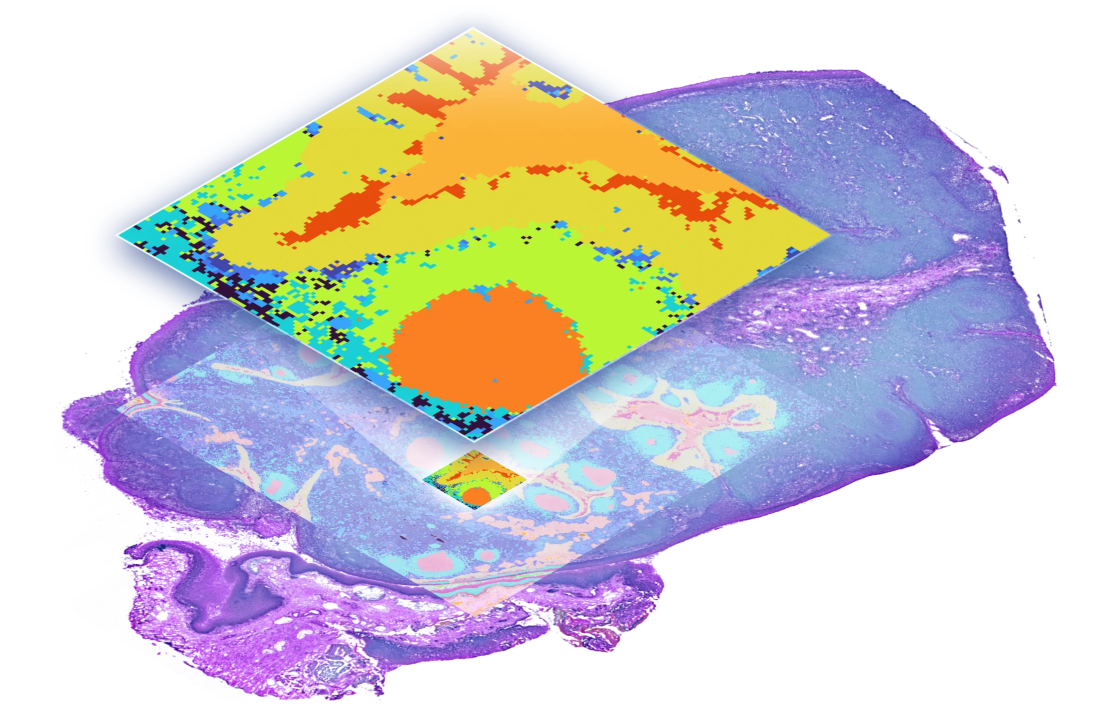
High-resolution, whole transcriptome spatial gene expression across species and tissue conditions
Visium CytAssist instrument
Enables a histology-friendly workflow and provides high spatial fidelity
Data analysis
Free, easy-to-use software for intuitive data visualization and exploration
Deeper spatial discovery
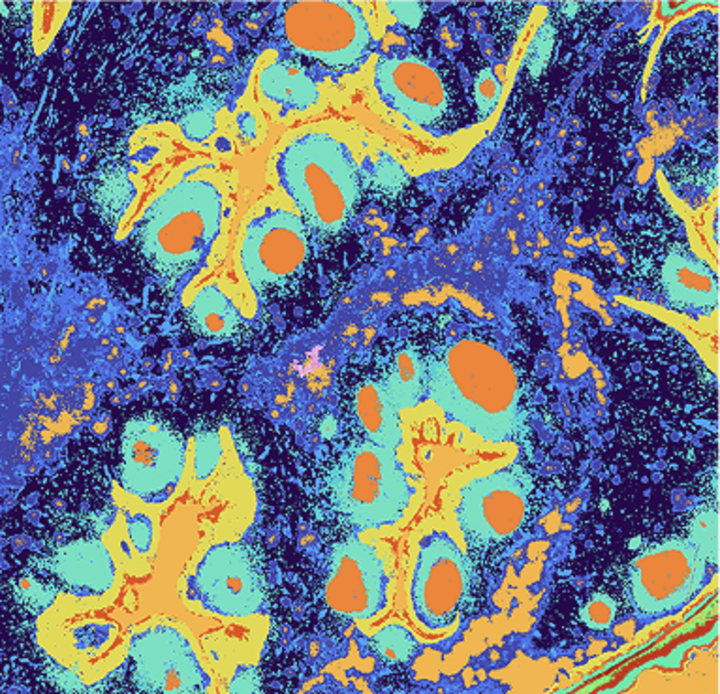
Species-agnostic, whole transcriptome spatial analysis at single cell–scale resolution with cell segmentation
De novo discovery, including feasibility for isoform, TCR/BCR, and SNV detection
High spatial fidelity and a streamlined, histology-friendly workflow powered by the Visium CytAssist instrument
Whole transcriptome spatial biology at the resolution you need
Visium HD WT Panel
Recommended for differential gene expression in human and mouse
Protein coding gene coverage: Optimized for archival and FFPE samples
Flexible tissue compatibility: Compatible with FFPE, fresh frozen, and fixed frozen tissues
Products
HD WT Panel Gene ExpressionNEW! Visium HD 3'
Recommended for expanded discovery applications
Whole transcriptome coverage: Delivers de novo discovery power
Explore across species: Tested in human, mouse, rat, pig, non-human primate and more.
Products
HD 3' Gene ExpressionUnmatched spatial data quality with a histology-friendly workflow

Ensure high spatial fidelity through precise transcript localization facilitated by the Visium CytAssist instrument
Leverage a standard histological workflow to section, stain, and image tissues directly on glass slides
Pre-screen and orient tissue sections to align the most desired region of tissue onto the Capture Area of the Visium slide
Accelerate spatial discovery with intuitive data exploration
Simple, fast analysis with Space Ranger
Upload FASTQ files to 10x Cloud Analysis or download our free software for local analysis
Process spatial sequencing data with imaging data in minutes with no command line required, including cell segmentation

Immediate insights with powerful data visualization
Visualize and explore thousands of spatially variable genes in a few clicks with Loupe Browser
Explore gene expression clusters in the tissue context to define cell types or gene signatures by region and morphology
Resources
Unlock the full spectrum of biology
| Platform | |||
| When to use | Comprehensive single cell dataIdeal for deep characterization of cell populations and states. | High-resolution spatial gene expressionUnderstand complex tissues, neighborhoods, and cell to cell interactions.
Integration with other spatial-omics, histology, and morphology. | |
| Why to use | Unbiased single cell discovery
High per-gene sensitivity | Unbiased spatial discovery | Targeted spatial exploration
High per-gene sensitivity |
| Applications | Whole transcriptome gene expression
Protein
TCR, BCR
CRISPR
ATAC | Whole transcriptome gene expression | Targeted gene expression (up to 5,000 genes) |
| Resolution | Single cell | Transcripts assigned to 2-µm areas | Single cell |
| Data readout | NGS-based | NGS-based | Imaging-based |
| Sample compatibility | Single cell or nuclei suspensions from fresh, frozen, or FFPE samples | FFPE
Fresh frozen
Fixed frozen | Fresh frozen
FFPE |