Spatial transcriptomics publications you’ll want to read: 2022 edition
You may remember this article from March of 2021, showcasing some of the latest and greatest research using Visium Spatial.
Since then, the spatial research landscape has changed quite a bit. Now, scientists are more aware of the value of spatial transcriptomics—and the increased number and diversity of published studies shows it. A bioRxiv search of “spatial transcriptomics” restricted to 2022 alone yields over 3,000 new results, suggesting a growing urgency to leverage a readout of spatially resolved gene expression for discovery and translational research.
Perhaps more impressively—and a testament to the labor and innovation of scientists spearheading the spatial movement—there are over 130 published studies in our publications database using Visium Spatial (including 11 from Cell, 7 from Nature, and 4 from Science).
Things have changed for us at 10x Genomics, too. We’ve introduced Visium Spatial for FFPE, which maps the whole transcriptome with morphological context in FFPE tissues, including precious clinical or archived samples. And we brought Visium CytAssist to customers this year, which expands sample access and simplifies the Visium workflow by facilitating the transfer of transcriptomic probes from standard glass slides to Visium slides.
Clearly, we’re not where we used to be. The future of spatial research is hurrying on as new studies and innovations build out technical capabilities and advance crucial insights into health and disease. Therefore, we thought it was necessary to pause and take stock of some of the incredible research that’s being done with spatial technology—to create a 2022 edition of the spatial transcriptomics publications you’ll want to read. In the following sections, explore a few select publications that reflect both the huge range of applications for spatial transcriptomics, as well as the amazing findings researchers are adding to their fields, from cancer biology to plant development.
Want to jump to topics within the blog? Explore pancreatic tumor heterogeneity, cardiac remodeling, T-cell activity in glioblastoma, a new model of photosynthesis, and orchid development.
Knowledge of tumor substructures promises to improve therapies
The Human Tumor Atlas Network (HTAN) has a lofty goal: “to construct 3-dimensional atlases of the dynamic cellular, morphological, and molecular features of human cancers as they evolve from precancerous lesions to advanced disease.” A team of scientists from Washington University in St. Louis, led by Dr. Daniel Zhou and Dr. Reyka Jayasinghe, recently added to the growing HTAN database with findings from their study of pancreatic ductal adenocarcinoma, a notoriously lethal cancer type with limited treatment options (1).
The team used single cell and single nucleus RNA-seq (sc/snRNA-seq) to profile 83 cancer samples from treatment-naïve patients and others undergoing a mix of treatment courses, including neoadjuvant targeting therapies and chemoradiation. They also generated matched spatial transcriptomics data for a subset of tumors, totaling 15 Visium slides. This multidimensional approach revealed cellular subpopulations with distinct histological features that characterize a transitional stage in cancer progression. In particular, the team observed the presence of acinar-to-ductal metaplasia cells—observed in animal models, but not characterized in human samples—and pancreatic intraepithelial neoplasia cells, which were confirmed to compose precursor lesions (1).
Integrated single cell and spatial data also pointed to a potential target for immune checkpoint blockade: the TIGIT–NECTIN axis. NECTIN4, encoding a surface ligand, was upregulated in tumor cells and showed spatial colocalization with tumor regions across samples. Likewise, TIGIT expression, encoding an immune receptor, colocalized near infiltrating lymphocyte regions in tumor sections. TIGIT interaction with NECTIN inactivates T-cell and natural killer cell effector functions. Thus, blocking NECTIN could support anti-tumor T-cell activity (1).
How the heart remodels after myocardial infarction
Though fears surrounding heart attacks tend to arise from the experience of the event itself, it’s what happens to the heart afterwards that continues to drive hospitalization and morbidity. One in five patients may be hospitalized with heart failure within 12 months following myocardial infarction (MI) as a result of left ventricular remodeling, structural and functional changes in the heart caused by loss of viable myocardium and a heightened inflammatory response, among other causes (2, 3).
Little is known about the mechanisms driving this remodeling event. Gaining a detailed understanding of the cellular and molecular players in the spatial context of the reorganizing heart tissue could unlock therapies to alleviate late-stage mortality. This motivation led a team of researchers from RWTH Aachen University and Heidelberg University in Germany, led by Drs. Christoph Kuppe, Ricardo Ramirez Flores, and Zhijian Li, to perform a comprehensive study of post-MI remodeling in the human heart using snRNA-seq, snATAC-seq (Assay for Transposase Accessible Chromatin), and spatial transcriptomics (4). Their approach allowed them to distinguish tissue structures of injury, repair, and remodeling, including a clear border zone between injured and uninjured cell types characterized by a gradient of ANKRD1 and NPPB gene expression (both associated with heart disease). Moreover, they observed that fibroblast-to-myofibroblast differentiation was a driver of heart tissue remodeling (4).
This spatial map of disease-specific cardiac cell states, along with their associated gene expression and gene-regulatory programs, provides a rich resource to address the underlying biology of cardiac remodeling for future therapeutic interventions.
T cells contribute to immunosuppression in glioblastoma
Cancer has many loopholes, including an ability to turn our own immune systems against us. While the degree of tumor immune infiltration can be associated with positive immunotherapeutic response, sometimes T cells in the tumor microenvironment can cause more harm than good. This “good” or “bad” effect is ultimately determined by the composition of T-cell populations, which is, in turn, highly influenced by crosstalk with cellular neighbors in the tumor microenvironment (5).
Researchers from the Medical Center – University of Freiburg in Germany sought to better understand this phenomenon in their study of glioblastoma, a malignant brain cancer that is highly resistant to immunotherapy (5).
First, using single cell gene expression on immune cells from 12 human tumor samples, the team discovered a subset of CD8+ T cells that exhibited classic exhaustion signatures correlated to IL-10 response, suggesting a role in T-cell dysfunction. Spatial transcriptomic analysis of tumor sections revealed that activated effector CD8+ T cells, exhausted CD8+ T cells, and CD4+ Th17-like T cells colocalized with tumor regions enriched for mesenchymal-like and astrocytic-like transcriptional signatures. Using an algorithm model to screen for cell types that are most likely to interact with each other and evoke the IL-10 response in T cells, they found that myeloid cells (macrophages and microglia cells) were the likely culprits for cellular crosstalk with infiltrating T cells in the mesenchymal-like tumor regions (5).
Understanding these cellular relationships represents an important step to boosting anti-tumor immunity in treatment programs for glioblastoma.
Spatial finds first evidence of an integrated photosynthesis mechanism
All of biological life is highly interconnected. That statement couldn’t be more evident than in the relationship between plant and human life. Plants—a food source, and a source of the essential ingredient to life, oxygen—keep the world spinning through photosynthesis, a series of chemical reactions that convert carbon dioxide into carbohydrates with the aid of sunlight and water.
There are different kinds of photosynthesis—C3 (the most common), C4, and CAM—that tend to occur in different plants based on their environmental conditions, such as temperature and access to water. Each process differs mostly in the mechanism of carbon dioxide fixation, but CAM photosynthesis is uniquely temporal: while C3 and C4 plants have open stomata (the respiratory center of leaves), CAM plants, adapted for hot, arid climates, keep their stomata closed during the day to retain water. In the cooler night, they then open to take in CO2 for fixation (6).
Some C4 plants are known to employ CAM photosynthetic reactions, but it’s been proposed that CAM activity must take place in distinct cell populations, such as specialized water storage cells (7). However, a team of researchers, led by Dr. Jose J. Moreno-Villena with corresponding author Dr. Haoran Zhou from Yale University, recently used spatial gene expression on Portulaca oleracea, a succulent weed, to discover that C4- and CAM-related genes operate in the same populations of photosynthetic mesophyll cells (7).
Understanding this previously uncharacterized biochemical pathway holds promise to generate both more photosynthetically efficient and drought-tolerant plants. (Read more about their study here.)
Beautiful organogenesis
Can we say we have a favorite Visium Spatial paper from 2022? It’s a hard choice—but these data visualizations of the developing orchid are persuasive.
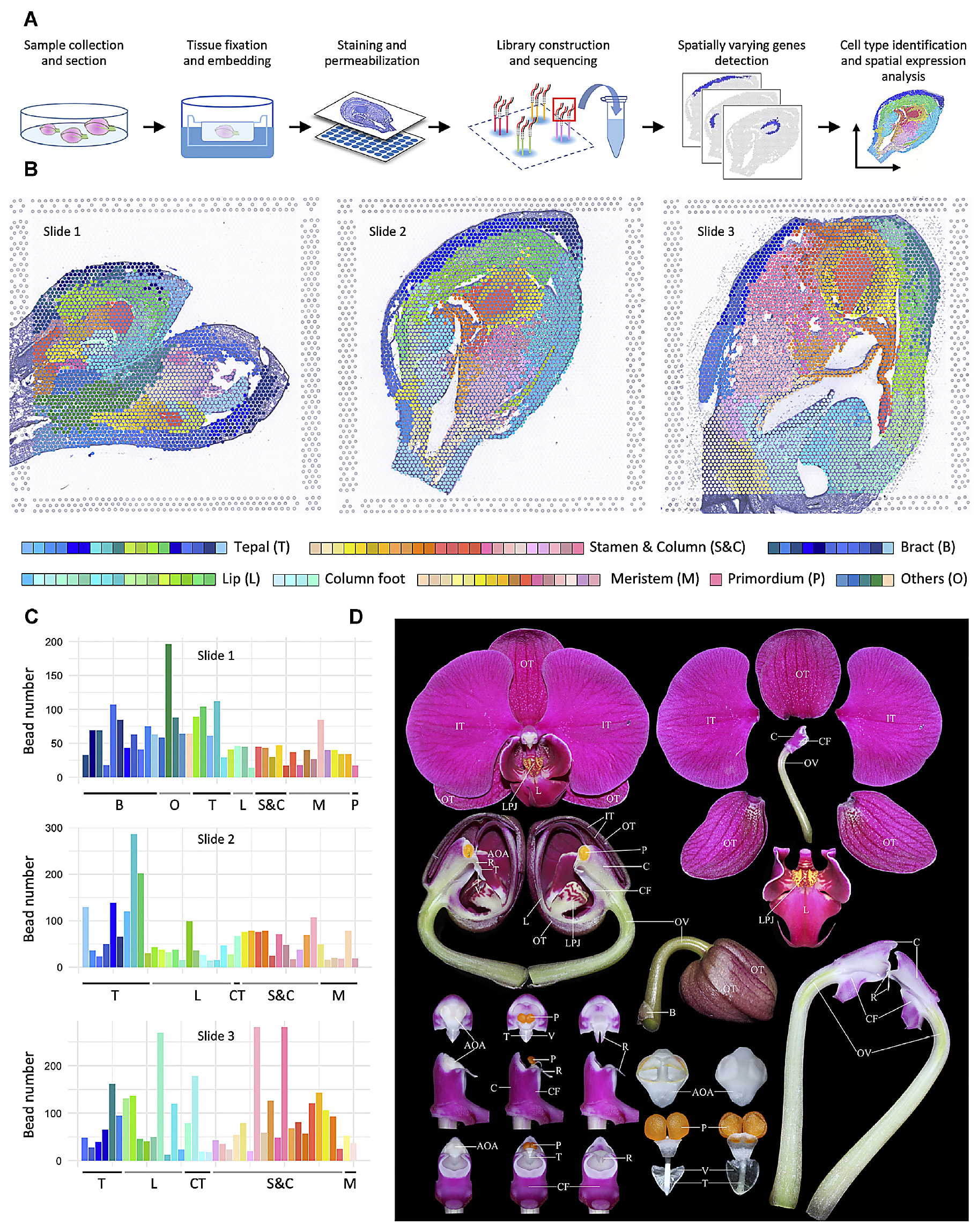
Led by Dr. Chang Liu, researchers from Fudan University and Jiangxi Agricultural University in China used spatial gene expression to create a comprehensive spatiotemporal map of floral organogenesis at multiple developmental stages in an orchid plant (8).
Their study resolved the preferential expression of thousands of genes in these distinct developmental stages, providing coverage of the continuous transcriptional events from initialization and identity determination to late morphogenesis, and a reference atlas for determining cell type–specific gene expression changes. This study models what’s possible with a whole transcriptome spatial readout of organ development in not only plants, but human and other animal tissues as well.
We’re always excited to share about the incredible research scientists are doing with 10x Genomics technology. Who knows what 2023 holds? As always, your work is what continues to drive the leading edge of spatial biology research—and inspires all that we do at 10x!
Want to discover more spatial research? Read about the first published study using our Visium for FFPE solution, exploring the role of tertiary lymphoid structures in cancer immunotherapy.
Find more resources for Visium Spatial Gene Expression →
References:
- Cui Zhou D, et al. Spatially restricted drivers and transitional cell populations cooperate with the microenvironment in untreated and chemo-resistant pancreatic cancer. Nat Genet 54: 1390–1405 (2022). doi: 10.1038/s41588-022-01157-1
- Niccoli G, et al. Optimized treatment of ST-elevation myocardial infarction. Circ Res 125: 245–258 (2019). doi: 10.1161/CIRCRESAHA.119.315344
- Abouzaki N and Abbate A. Causes and prevention of ventricular remodeling after MI. FACC Expert Analysis (2016).
- Kuppe C, et al. Spatial multi-omic map of human myocardial infarction. Nature 608: 766–777 (2022). doi: 10.1038/s41586-022-05060-x
- Ravi VM, et al. T-cell dysfunction in the glioblastoma microenvironment is mediated by myeloid cells releasing interleukin-10. Nat Commun 13(1) (2022). doi: 10.1038/s41467-022-28523-1
- What is the difference between C3 C4 and CAM photosynthesis? PEDIAA (2020).
- Moreno-Villena J, et al. Spatial resolution of an integrated C4+CAM photosynthetic metabolism. Sci Adv 8: eabn2349 (2022). doi: 10.1126/sciadv.abn2349
- Liu C, et al. A spatiotemporal atlas of organogenesis in the development of orchid flowers. Nuc Acids Res gkac773 (2022). doi: 10.1093/nar/gkac773
