Strengthening the core: collaboration when tissue meets genomics at Newcastle University
What do pathologists and tissue microscopy experts say to genomics experts and bioinformaticians? At Newcastle University, they say, “Hey, let’s work together.” As genomic approaches become staples of biological research, scientific partnerships like these will help power new discoveries. For scientists at Newcastle, collaboration was forged by each group’s common ties to Visium Spatial Gene Expression, a solution offered by 10x Genomics that enables unbiased characterization of spatially resolved whole transcriptome gene expression in thousands of spots across a section of tissue on a tissue slide.

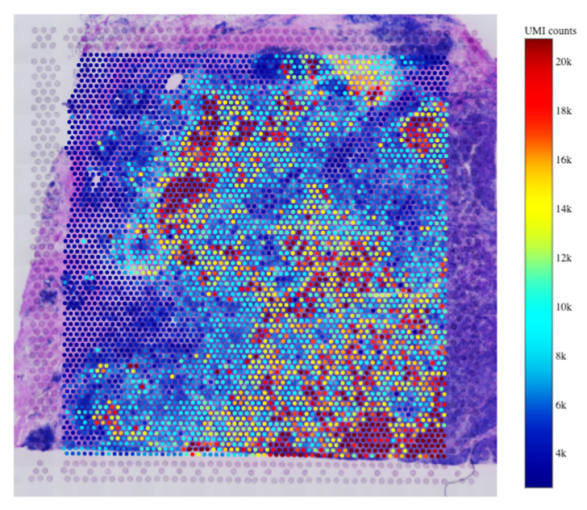
Newcastle University is a Certified Service Provider (CSP) for 10x Genomics, specializing in the Visium Spatial Gene Expression workflow. This means that researchers across the world can leverage the expertise of a team of certified scientists at Newcastle to conduct the spatial gene expression profiling technique on their tissue samples. Resident experts for each aspect of the spatial gene expression workflow, from sample preparation to data analysis and visualization, come together to perform a complete experiment. This SpaRTAN service, short for Spatially Resolved Transcriptomics At Newcastle, demonstrates powerful scientific collaboration that translates to other researchers looking to do spatial gene expression profiling. Whether leveraging a Certified Service Provider, or working with the resources available at their own institution or university, there are ways for researchers to finish their experiments efficiently, without needing to become an expert in every aspect of the process.
Join us as we follow a tissue sample through the SpaRTAN service and hear from sample preparation, cryosectioning, microscopy, genomics, and bioinformatics experts at Newcastle University as they discuss their collaboration. Find out how core facilities can come together to accelerate spatial gene expression experiments, and explore tips for optimized collaboration to leverage for your own team science.
Bringing experts together at every stage
The Visium Spatial Gene Expression workflow begins with fresh or fresh-frozen tissue. We spoke to Dr. Steve Lisgo, whose team consists of experts in tissue optimization, cryosectioning, and slide preparation at the university’s Human Developmental Biology Resource (HDBR), to find out more about this step. In addition to serving as a human embryonic and fetal tissue bank, the HDBR gives researchers access to gene expression services using immunohistochemistry and in situ hybridization techniques. “Spatial transcriptomics seemed like a logical extension to these services,” shared Dr. Lisgo.
Within the spatial workflow, Dr. Lisgo said, “the HDBR is responsible for the sample prep and the experimental work up to the library prep stage. We have a lot of histology and cryosectioning experience, as we do these kinds of things routinely within the tissue bank.” Their process includes a QC check “for RNA quality and histological appearance. The sample is then embedded in the OCT and frozen in an isopentane bath. The tissue is then taken through the standard Visium protocol.”
Dr. Lisgo also spoke to the holistic process and the benefits of collaboration across core labs at Newcastle. “We were fortunate that [team members in our genomics and bioinformatics cores] have been working together providing single cell services for a couple of years, so we knew that the transition between sequencing and data analysis would be fairly straight forward...Tapping into each other’s experience is definitely a better approach than starting out on your own.”

The next steps in the Visium workflow take a researcher into the microscope room, where image capture is performed to enable histological analysis of each tissue section. At Newcastle, that researcher is Dr. Alex Laude. He described the capabilities of the Bio-Imaging Unit, “a microscopy core facility, providing access to bright field, widefield fluorescence, confocal, super-resolution, high-content, and multi-photon imaging technologies. Unit staff advise on all aspects of the imaging workflow, from experimental design to image analysis.”
Dr. Laude offered his insights about how to optimize the imaging component of the SpaRTAN workflow. “It requires plenty of meetings, discussions, and trial runs to make sure that the imaging resolution is sufficient and the orientation and alignment of the tissue is correct.” Dr. Laude also noted the key to working well with their spaRTAN partners is “effective and regular communication so that all parts of the spaRTAN workflow align and come together at the right time.”
Following this imaging step, Visium Spatial Gene Expression slides are taken through permeabilization steps that release mRNA from the tissue cells. mRNA products are barcoded with a unique spatial barcode to map them back to a single tissue spot. Then they’re processed further in preparation for library construction. Dr. Jonathan Coxhead’s team members take the cDNA forward for library preparation and subsequent sequencing steps in the Genomics Core Facility (GCF) at Newcastle University. “We’re a next-generation sequencing service provider that specializes in single cell genomics. We have been working with single cell technologies since 2015 and have been working with 10x Genomics for almost 3 years. We also offer qPCR, ddPCR and high-throughput exome, genome, and RNA-seq services alongside our single cell applications. In addition to being a university core facility, we carry out contract work for academia, industry, and health care sectors.”
Dr. Coxhead provided some thoughts about how the core labs at Newcastle have learned to work together, and how their experience can inform other groups and researchers looking to collaborate to run their experiments. “We are all specialists in key areas, and it was just a case of working out the best handover, and safe stopping points within the workflow...Having communities that already work together is the ideal situation, as well as having accompanying technologies within fairly close proximity to each other. Alternatively, it is helpful to identify someone who’s worked on similar workflows before, like single cell genomics, who can provide oversight to bring teams together.”
As they prioritized developing smooth handoffs and processes, from tissue input to data output, for the Visium Spatial Gene Expression workflow, Dr. Coxhead remarked that the Newcastle core labs were increasingly able to meet scientists’ needs for their services. “...We’re gaining experience all the time, which in turn benefits our end users as we’re able to offer advice on how to handle different tissue types and help optimize experimental design.”

We spoke to Dr. Rachel Queen of the Bioinformatics Support Unit at Newcastle University to understand the final steps of the SpaRTAN service, including analysis and visualization of spatial gene expression data, which is layered on top of the histological images of the tissue sections captured earlier in the workflow. Dr. Queen described the capabilities of her core facility: “We provide analysis and experimental design for high-throughput experiments. For Visium specifically, the bioinformatics core is responsible for analyzing the data, creating an analysis report, and the transfer for the data at the end of the project.” These steps leverage 10x Genomics’ software tool Space Ranger and other tools Dr. Queen has worked with during her years of providing support for single cell experiments. “I process the samples using Space Ranger and then do a combined analysis of adjacent sections in R. I wrote an R package called Spaniel for plotting data onto the histological image. I also use a range of single cell R packages from Bioconductor and the Seurat R package.”

Better science together
Each core lab brings their own area of expertise to the Visium Spatial Gene Expression workflow. In coming together, they strengthen their experiments and improve the services they provide to other researchers. We appreciate these insights from our partners at Newcastle University, and we’re inspired by their demonstration of the value of collaboration, where individual strengths shine and drive better, more efficient science.
To learn more about the services provided by Newcastle University, visit their website.
Are you looking to start profiling spatial gene expression in your tissue samples? Explore Visium Spatial Gene Expression here.
This article contains a discussion of research and processes conducted by scientists at Spatially Resolved Transcriptomics At Newcastle (SpaRTAN), Newcastle University. View and opinions do not constitute endorsement or promotion of 10x Genomics, Inc. or any of its products.
