Tracing T cells in brain metastases with combined spatial and T-cell receptor sequencing
The Innovator Blog Series celebrates research conducted by 10x Genomics customers who have demonstrated scientific ingenuity by adapting Chromium Single Cell or Visium Spatial sequencing-based assays. These innovations are distinguished by their originality and potential impact on scientific discovery, including providing access to novel analytes, advancing multiomic analysis techniques, and demonstrating critical applications to human health and disease. These techniques are customer developed, meaning they are not officially supported by 10x Genomics.
In this article, we highlight a method for combining spatial gene expression and T-cell receptor sequencing from a research team at the Emory University School of Medicine in Atlanta, GA. Learn how this technique works, and explore the insights it is providing into immune localization in the tumor microenvironment of brain metastases.
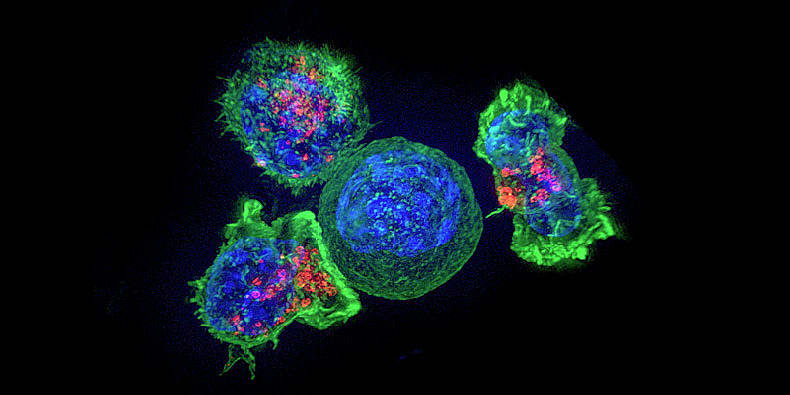
Why T-cell infiltration matters in metastases
It’s the news that no cancer patient ever wants to hear. Metastasis—when cancer cells from a primary tumor spread to other distant parts of the body, depositing cancer colonies in previously untouched organs where uncontrolled cellular growth continues to cause harm. Having a single tumor is scary enough, but the greater physical and emotional toll may come for patients, families, and healthcare providers when treatments just can’t seem to keep cancer growth in check.
In primary tumors and metastases alike, prognosis is highly dependent on immune activity in the tumor microenvironment. For example, in their 2017 review in the British Journal of Cancer, Barnes and Amir noted that “a ‘pro-inflammatory’ tumor microenvironment and infiltrating CD8-expressing lymphocytes are associated with improved clinical outcomes in a broad range of tumor types” (1). This same effect is noted in metastases: “Tumor infiltrating lymphocytes (TILs) in brain metastases are positively correlated to the prognosis... [one] study analyzing 25 pairs of primary NSCLC (non-small cell lung cancer) specimens and brain metastases of NSCLC found that the overall density of CD8+ T cells in the parenchyma of brain metastases is higher than that in primary foci, and patients with a lower number of CD8+ TILs in the matrix present a worse prognosis” (2).
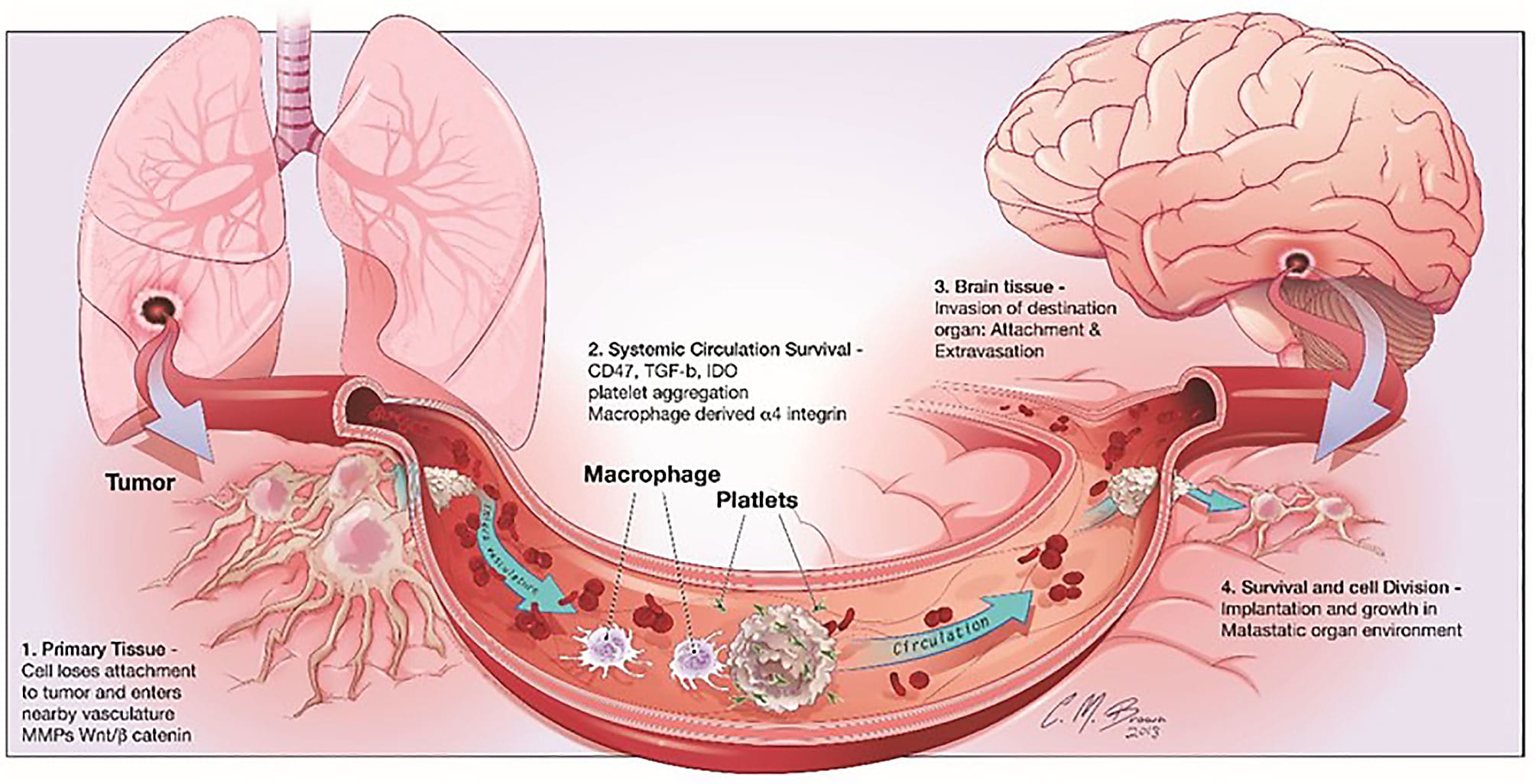
But, as Barnes and Amir qualify, not all T cells have the same positive effect in the tumor microenvironment. “The inhibitory function of other immune cells, for example, myeloid-derived suppressor cells and regulatory T cells (Tregs) appear to have a major role in disrupting the capacity for the immune control of cancers and are therefore associated with worse outcome” (1). Other recent discoveries point to models of cancer evolution and therapeutic resistance hinging on the composition of the tumor-immune infiltrate. Using single cell RNA-sequencing to study the tumor microenvironment of uveal melanoma (UM), scientists from the Comprehensive Cancer Center of the University of Miami discovered specific immune cell populations that were not previously associated with UM. This included T cells with an alternative checkpoint marker, LAG3, that was more broadly expressed in UM tumors than the traditional CTLA4 and PD1 markers (3).
Distinguishing T-cell functional states, antigen specificity, and other overall characteristics in the tumor microenvironment (TME) is therefore essential to understanding cancer prognosis, including why some cancers don’t respond to treatment and metastasize. But the molecular characteristics of TILs only represent one layer of complexity; each cancer type, even each specific tumor, has unique qualities that can influence both the composition and spatial organization of the immune microenvironment. In other words, the extent of immune cell infiltration in a tumor also informs prognosis.
In a study of triple-negative breast cancer, Visium Spatial Gene Expression data revealed a ring of exhausted T cells around the tumor mass in a tissue section (4). Importantly, spatially resolved gene expression could provide a view of both the identity of infiltrating immune cells and extent of penetration into the cancerous tissue at a higher resolution than that provided by pathological annotation alone—in this case, suggesting the infiltrating T-cell population had limited anti-tumor efficacy.
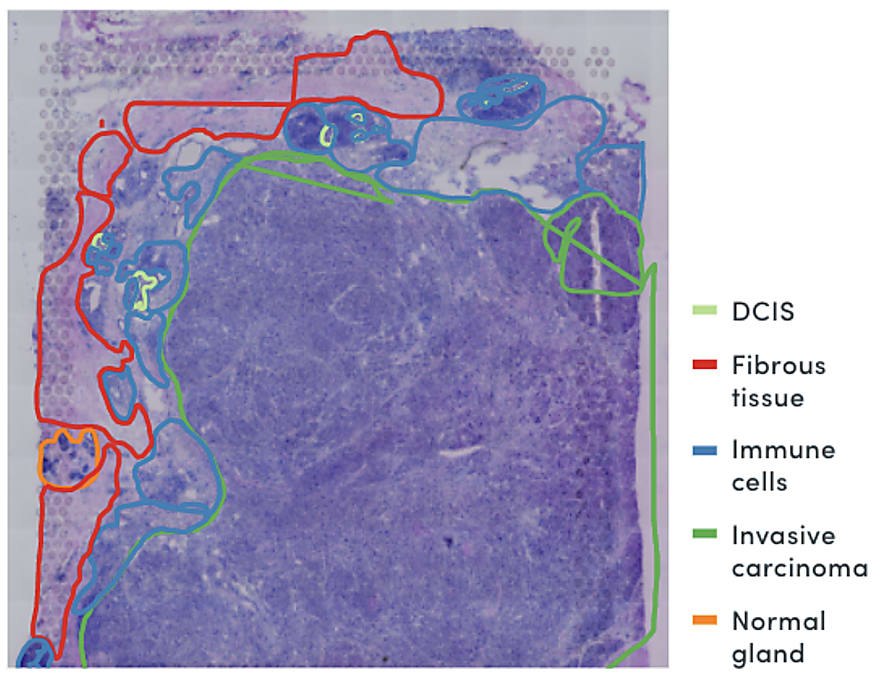
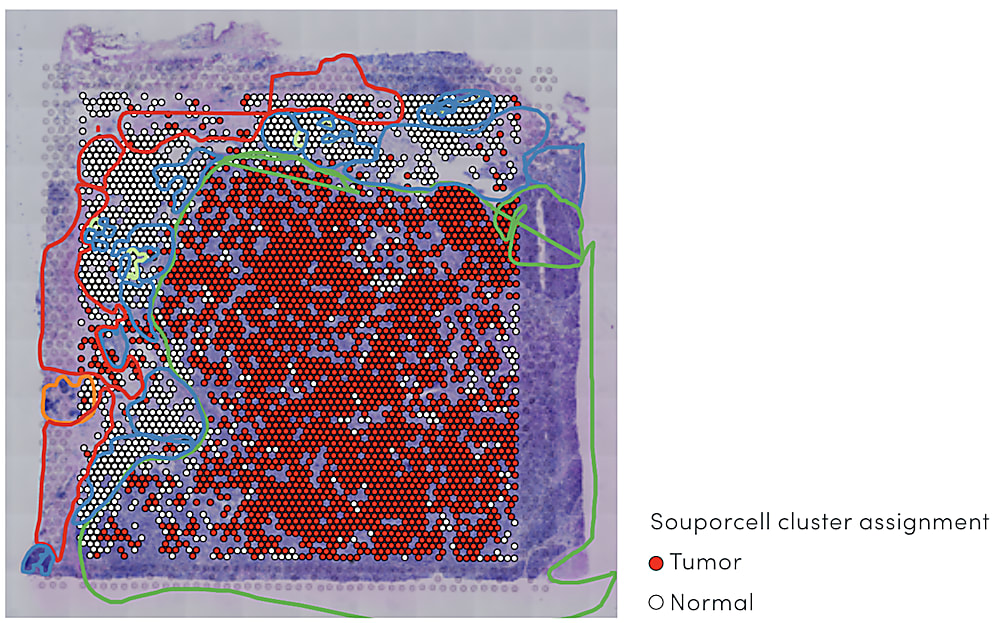
Combining spatial gene expression and T-cell receptor sequencing
These complex factors at play in the immune response to cancer point to the need for a comprehensive view of the TME that unifies multidimensional readouts, including single cell and spatial annotation of T-cell composition and infiltration. In their recent bioRxiv preprint, Dr. William Hudson, Dr. Lisa Sudmeier, and their colleagues at the Emory University School of Medicine in Atlanta, GA reported a method to define T-cell receptor (TCR) sequences through Visium Spatial Gene Expression technology from 10x Genomics. As the authors noted, T-cell receptor sequences “determine lymphocyte antigen specificities and can serve as a barcode for lymphocyte clones,” a key readout of T-cell identity and function. In this way, the method would allow them to simultaneously visualize T-cell receptor clonotypes and overall gene expression within tissue samples in a single assay (5).
To accomplish this, the authors modified the standard cDNA library preparation steps conducted in the Visium assay. They added primers to the 5’ end of TCR sequences to simultaneously capture the variable CDR3 region of the TCR and the spatial barcodes containing the location information of the transcript within the tissue section. They also modified the library cleanup steps: the standard Visium assay requires fragmentation of the cDNA library, but in this case they cleaned the library through bead purification to retain the linkage between TCR and spatial barcode sequences (5).
Read more about their protocol modifications here →
Looking at T-cell infiltration in brain metastases from multiple angles
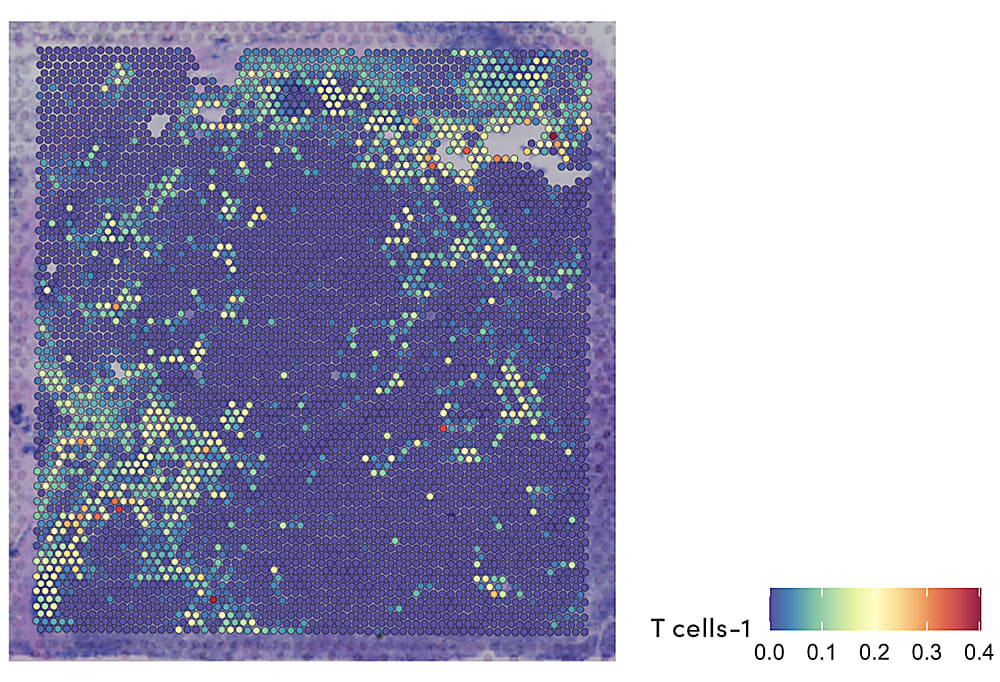
Using this method to obtain TCR sequences from spatial transcriptomics data, the research team performed a detailed characterization of infiltrating CD8+ T cells and their surrounding tumor microenvironment in brain metastases surgically removed from 31 patients. They identified four main transcriptional populations among PD1+ CD8+ T cells, including dividing cells, terminally differentiated cells, and two clusters with similar features to exhausted progenitor cells. A spatially resolved readout of the TCR sequences and gene expression also showed that each population was localized to a specific region of the brain metastasis tumor microenvironment, suggesting this diverse mosaic of CD8+ tumor-infiltrating T cells organized according to antigen specificity (6).
Explore more of their findings and data here →
In addition to guiding potential immunotherapeutic strategies for brain metastases, these findings demonstrate the value of a systems approach to biology—where cellular heterogeneity is unveiled through information obtained from more than a single parameter, and built upon the insights of a multiomic view of biology. Here, T-cell transcriptional signatures, spatial localization, and TCR–antigen specificity came together to build out the most comprehensive understanding of adaptive immune processes. With access to these analytes, scientists can continue to lead the way in resolving the complex immune activity in the tumor microenvironment and the multidimensional factors that drive cancer prognosis.
References:
- Barnes T and Amir E. HYPE or HOPE: the prognostic value of infiltrating immune cells in cancer. BJC 117: 451–460 (2017). doi: 10.1038/bjc.2017.220
- Luo L, et al.The Immune microenvironment in brain metastases of non-small cell lung cancer. Front Oncol 11: 698844 (2021). doi: 10.3389/fonc.2021.698844
- GenomeWeb and 10x Genomics. White Paper. Using Single-Cell Multiomics to Reveal New Insights Into Uveal Melanoma Evolution and Therapeutic Resistance.
- 10x Genomics. Application Note. Spatially Resolved Heterogeneity of Triple Negative Breast Cancer.
- Hudson W and Sudmeier L. Localization of T cell clonotypes using spatial transcriptomics. bioRxiv. Posted August 5, 2021. doi: 10.1101/2021.08.03.454999
- Sudmeier L, et al. The CD8+ T cell landscape of human brain metastases. bioRxiv. Posted August 5, 2021. doi: 10.1101/2021.08.03.455000
