Uniting powerful tools for tissue analysis: mRNA and protein co-detection
Achieve comprehensive tissue phenotyping with spatially resolved whole transcriptome and immunofluorescence protein co-detection. Learn more about our new protocol, Visium Spatial Gene Expression with Immunofluorescence, in this upcoming webinar.
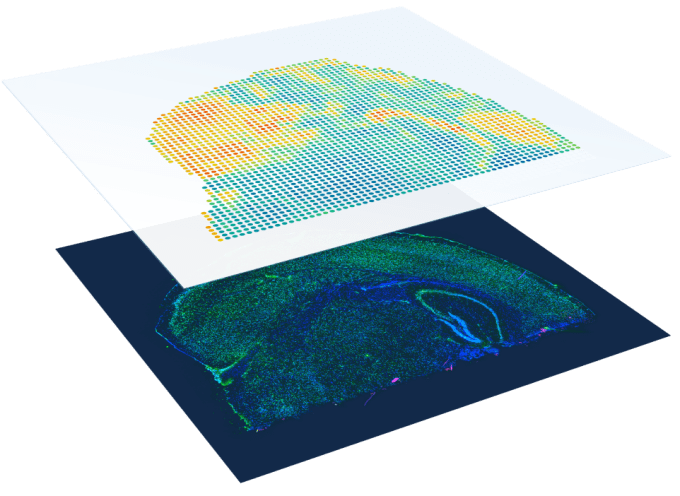
Pathologists have developed the incredible ability to visually annotate the complex biology captured on a tissue slide. Leveraging the traditional H&E stain and a microscope, they can distinguish and classify areas of disease within tissue, and reach crucial diagnosis conclusions.
Due to the complexity of tissue biology, and the constant need for greater resolution to study that complexity, scientists sought to develop molecular tools for spatial tissue analysis. They found that fluorescently labeled antibodies could localize a selection of known protein targets, providing a picture of cellular localization and post-translational gene expression in the tissue context. Moreover, they looked to merge genomics and pathology, providing tools that offer spatially resolved molecular annotation of gene expression and pathological gene signatures.
Now, with tools like Visium Spatial Gene Expression, researchers can access a multidimensional profile of the biology of a complex tissue section, measuring and mapping spatial gene expression while maintaining a clear view of the morphological context of the tissue. While this provides powerful molecular resolution for pathological annotation, it also provides unbiased discovery potential as scientists can explore the tissue context and uncover the influence of spatial architecture on gene expression, cellular phenotypes, the progression of diseases like cancer or neurodegeneration, and discover novel biomarkers.
But with biology there is always more to uncover. And, as the Visium platform demonstrates, there is value in uniting powerful methods and tools for histology and tissue analysis. Now, researchers can pull back another layer of tissue complexity using the Visium Spatial Gene Expression with Immunofluorescence workflow. They can leverage their own validated fluorophore-tagged antibodies for immunofluorescence stains of 2–3 proteins, but subsequently gain a full view of the mRNA expression across an entire section of tissue with the Visium protocol.
There is amazing potential across research areas with these combined technologies. For example, researchers can:
- Spatially map cell types of interest in heterogeneous tissue samples while accessing their full gene expression profiles
- Refine anatomical tissue and cell atlas spatial maps through a more comprehensive characterization of cell types at both the protein and transcript level
- Determine the activation or exhaustion states of infiltrating immune cells in the tumor microenvironment by tracking the cellular sources of secreted proteins
- Uncover alterations in the brain microenvironment caused by neuropathology, and determine the relationship between pathological protein aggregates and neurotoxicity
- Identify new tumor biomarkers and drug targets with cell type specificity
Explore the capabilities of Visium with Immunofluorescence by watching our upcoming webinar, Introducing Visium Spatial Gene Expression with Immunofluorescence: Gain a New Perspective on Tissue Complexity. Learn how to combine these powerful tools to capture multiple analytes in the same tissue sample, and retain current histological laboratory methods, as 10x-perts walk through the full workflow. Watch an interactive demonstration of 10x Genomics software tools on cancer and brain tissue samples to learn how you can derive meaningful biological results from your Visium with Immunofluorescence datasets.
Check out these snapshots of our software tutorial:
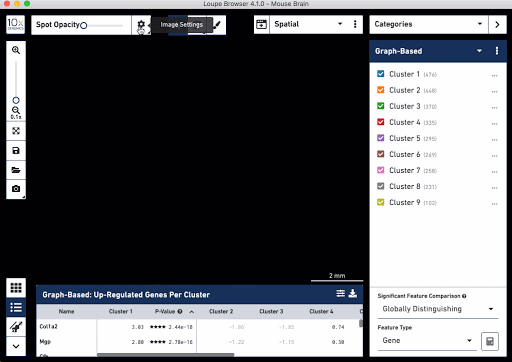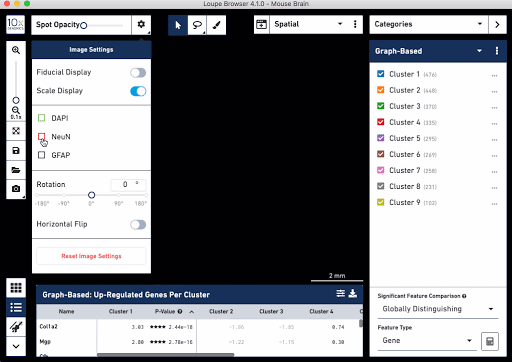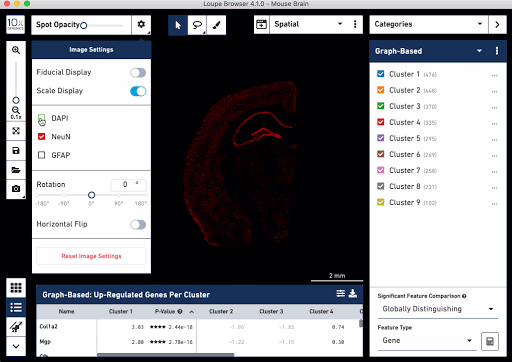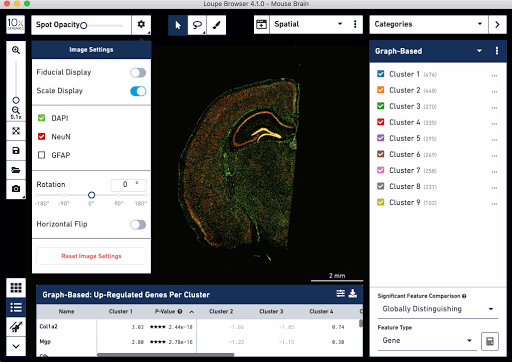
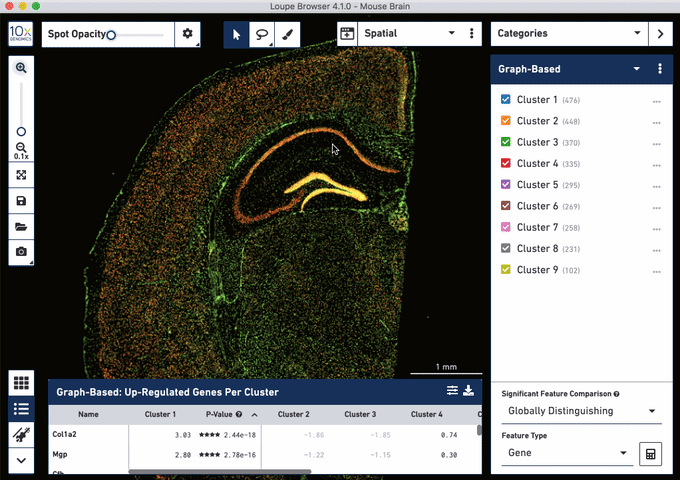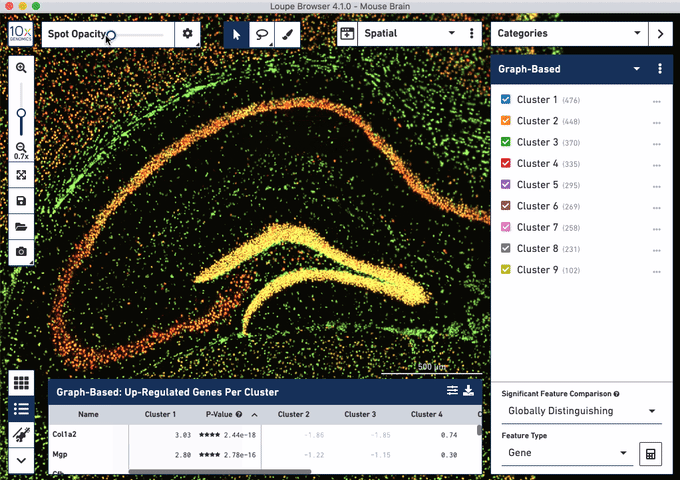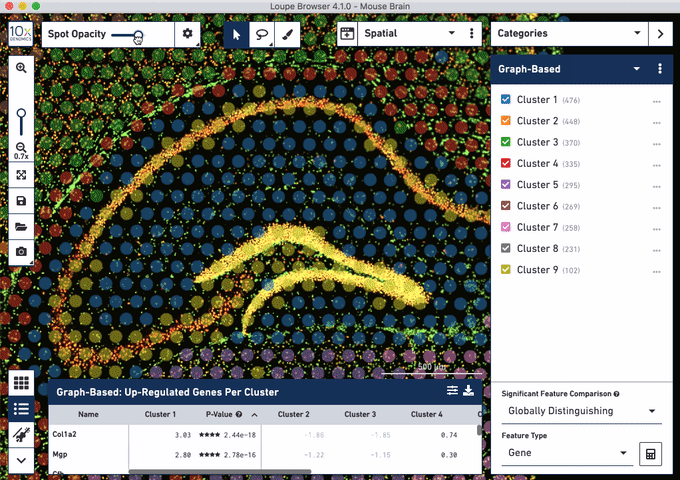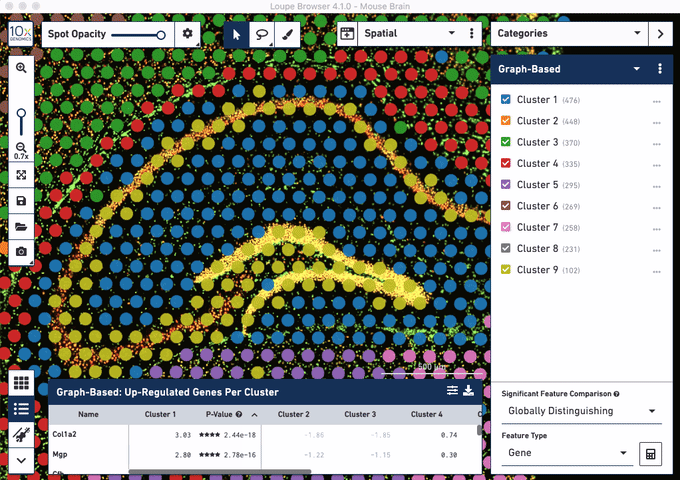
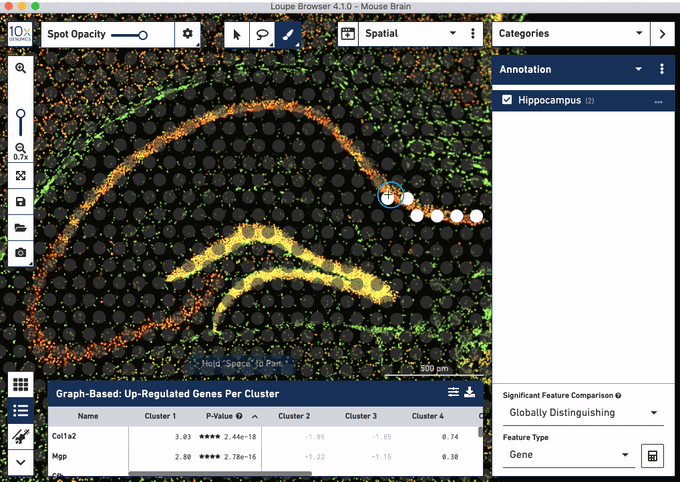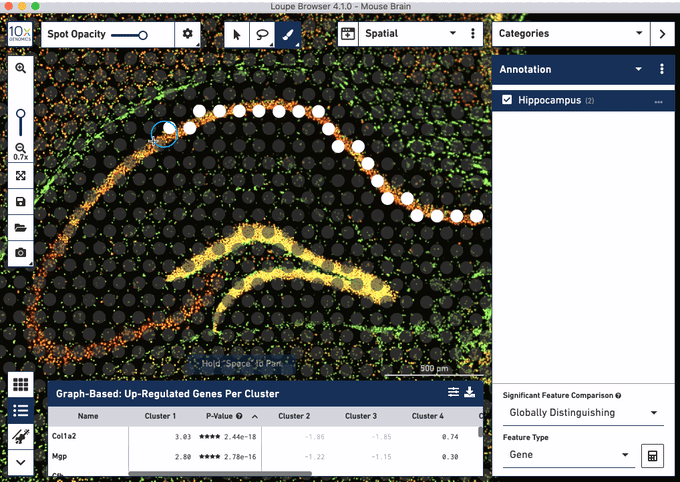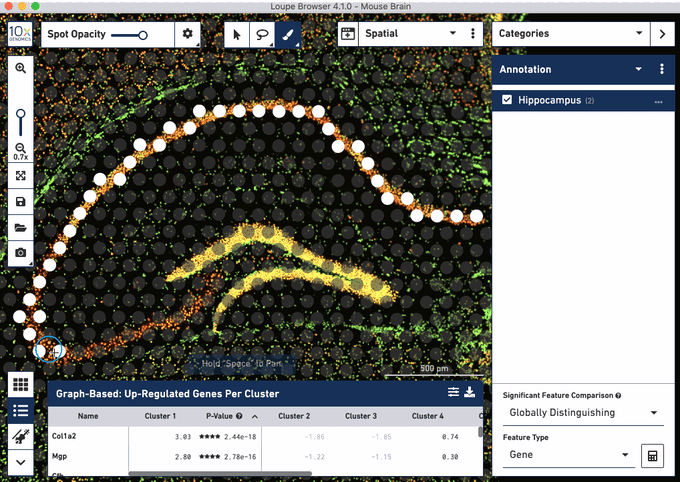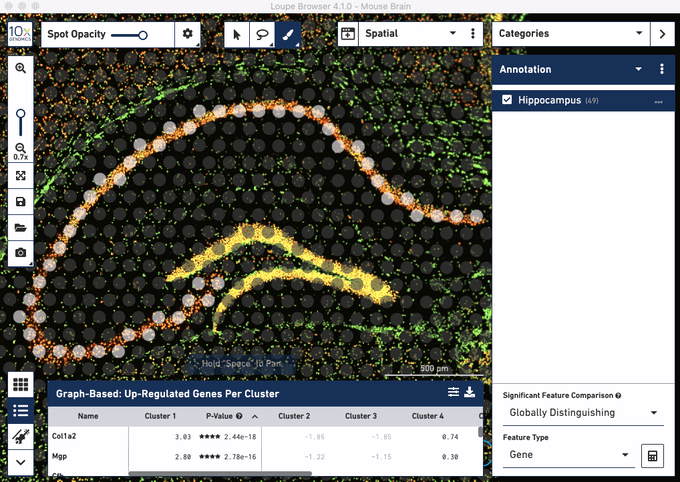
Explore the Visium with Immunofluorescence workflow and watch the full software tutorial in our upcoming webinar on July 7 (9:00 AM PDT/6:00 PM CEST), and on July 10 (10:00 AM SGT). Register here for PDT, and here for SGT.
Learn more about Visium with Immunofluorescence →
