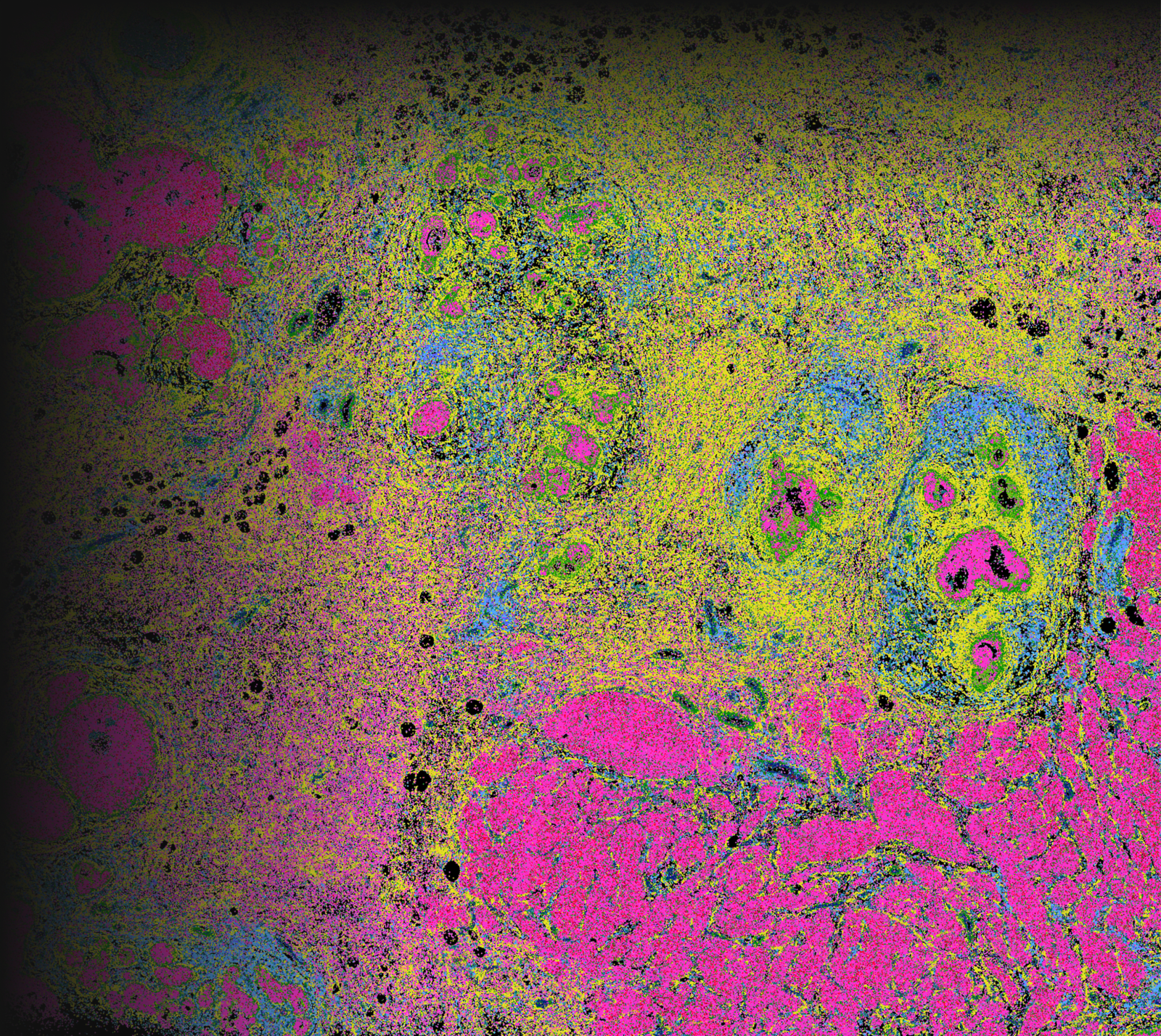
これは、Xeniumの開発モデルから得られたXenium In Situデータのプレビューです。
Xenium Analyzerの改良モデルで得られた新たなデータは、www.10xgenomics.com/datasetsから取得できます。
主な解析結果
- 生物種および組織ヒト乳腺
- 保存方法FFPE
- 使用した既製パネルXenium ヒト乳がん遺伝子発現パネル(280個の遺伝子)
- カスタム遺伝子の数33
- 組織面積0.4 cm²
- 検出された細胞数*167,780
- 検出された転写産物の総数*34,442,716
- 細胞あたりの転写産物数の中央値*166
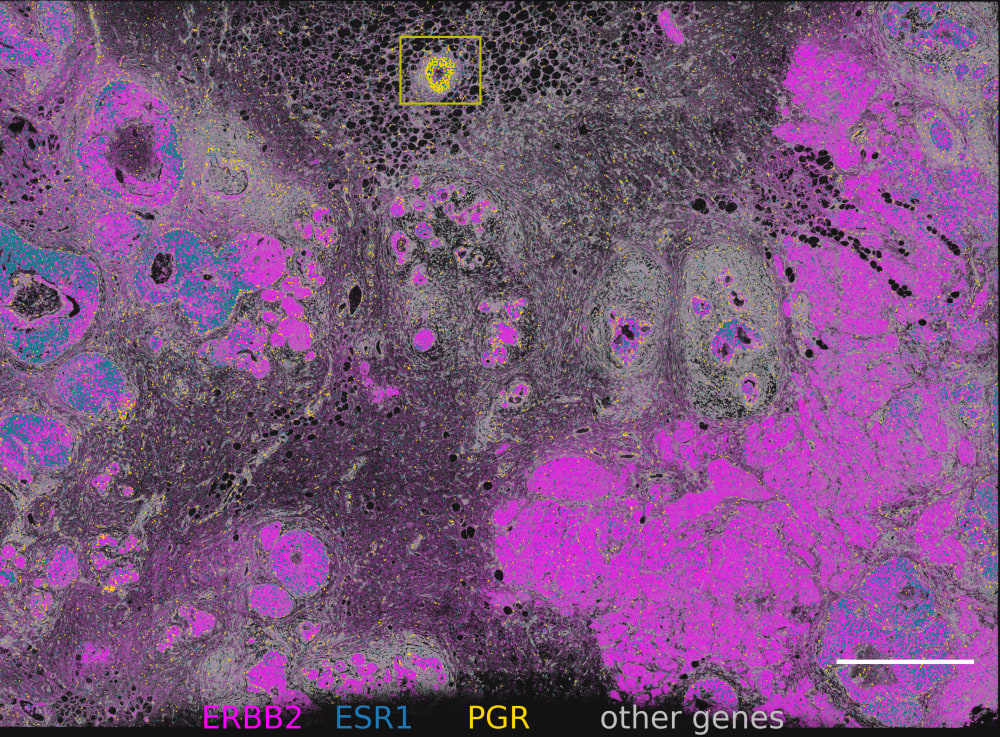
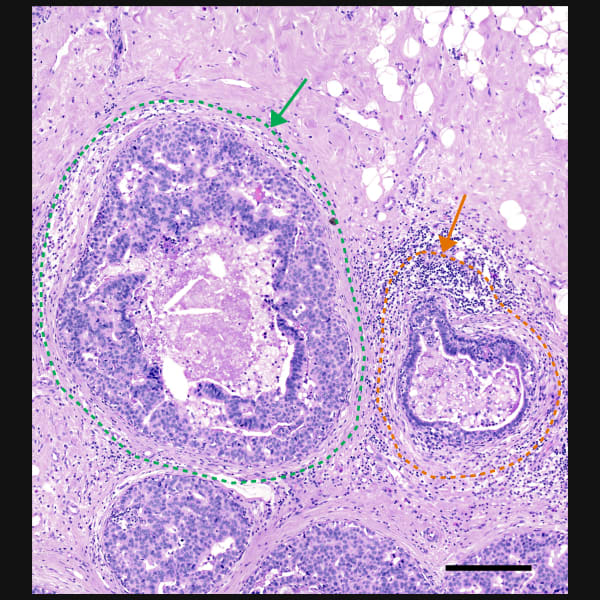
Xeniumで細胞種の不均一性を探索し、同じ切片をHEで染色して確認
細胞種と遺伝子特性の空間的構成を視覚化することで異なる組織マクロ構造を明らかにし、腫瘍領域内の細胞や分子に大きな不均一性があることを把握します。
Xeniumはヒト乳腺の主要な細胞種を同定します。2つの例で示すように、腫瘍サブタイプの不均一性および腫瘍上皮細胞周囲の微小環境を研究するためには空間情報が必要です。Xeniumのラン後に取得するH&E染色データは、転写産物情報を解釈し検証する際に役立ちます。
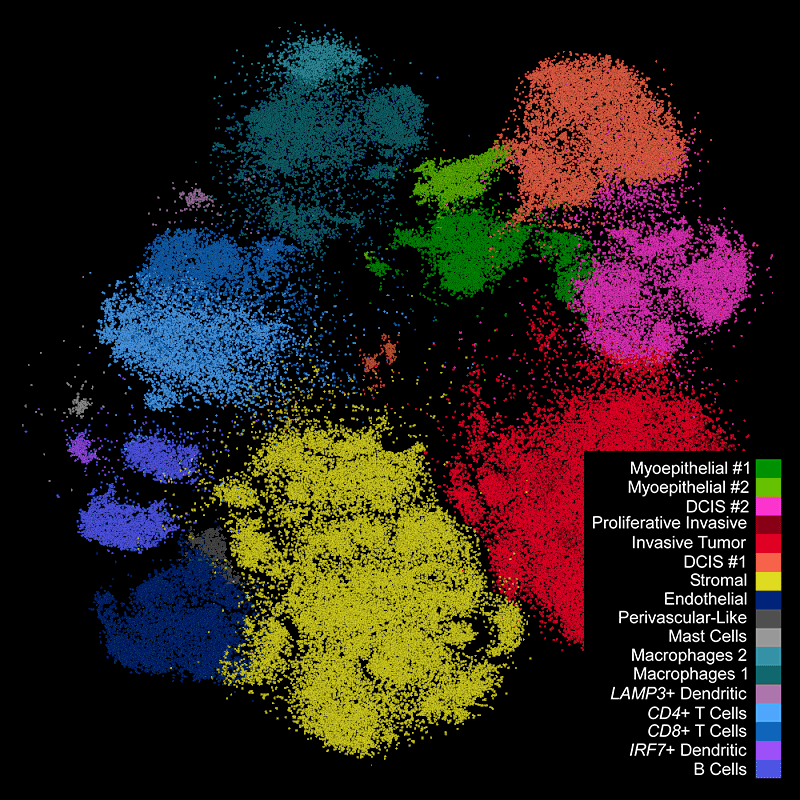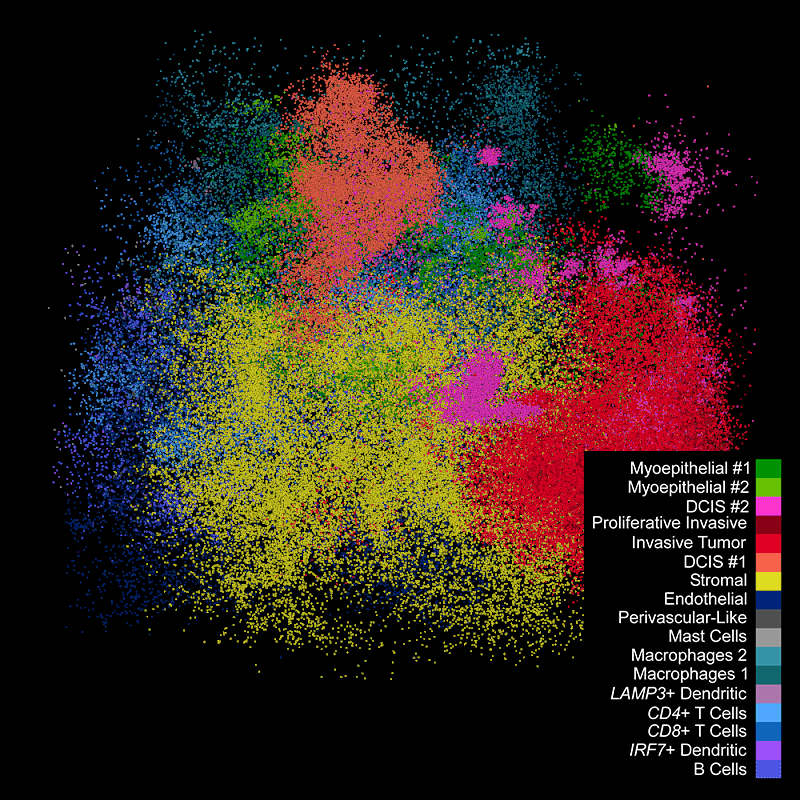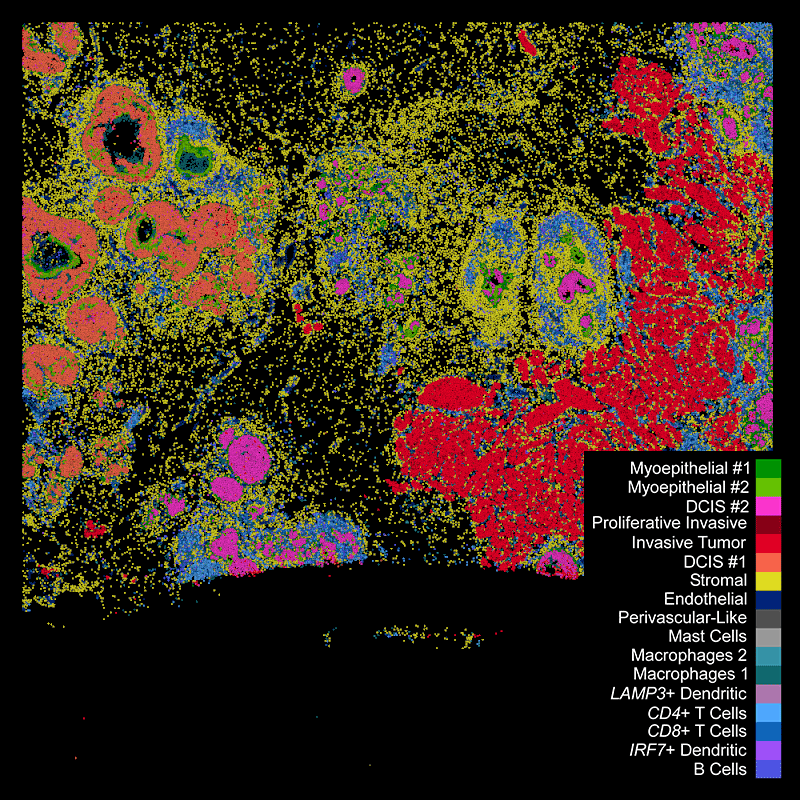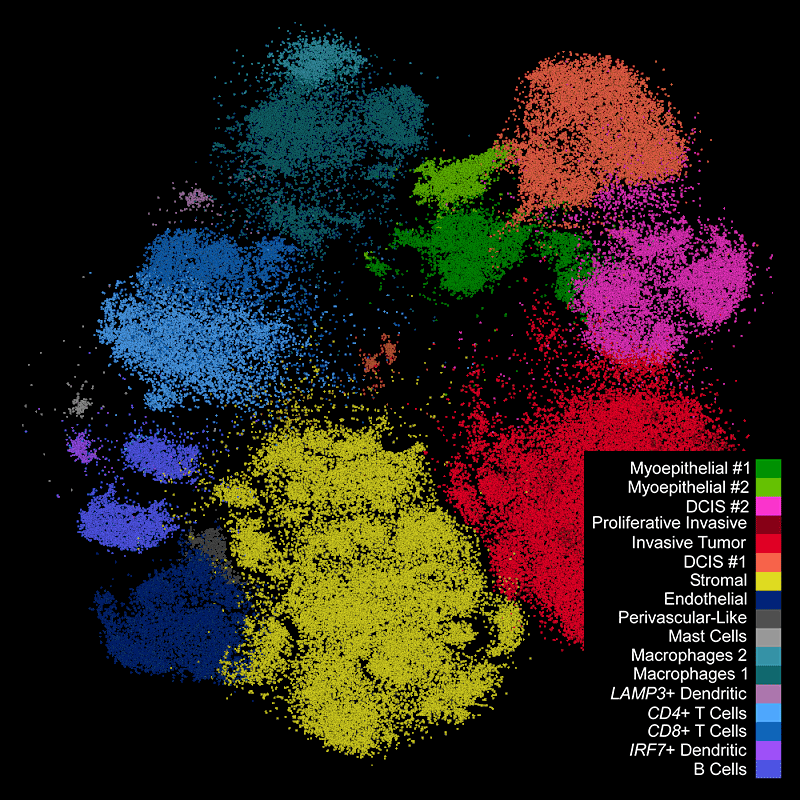
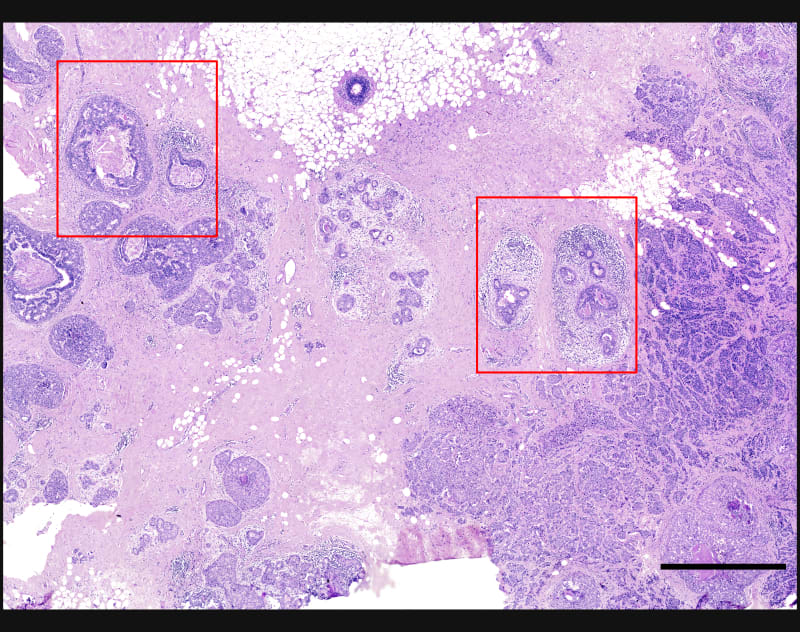
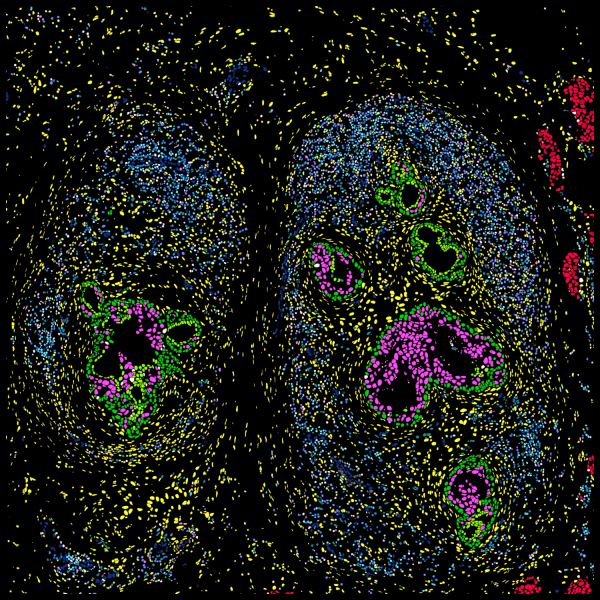
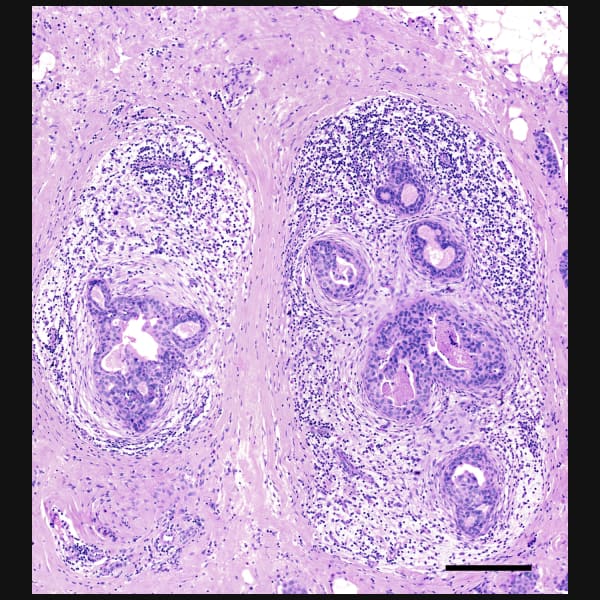

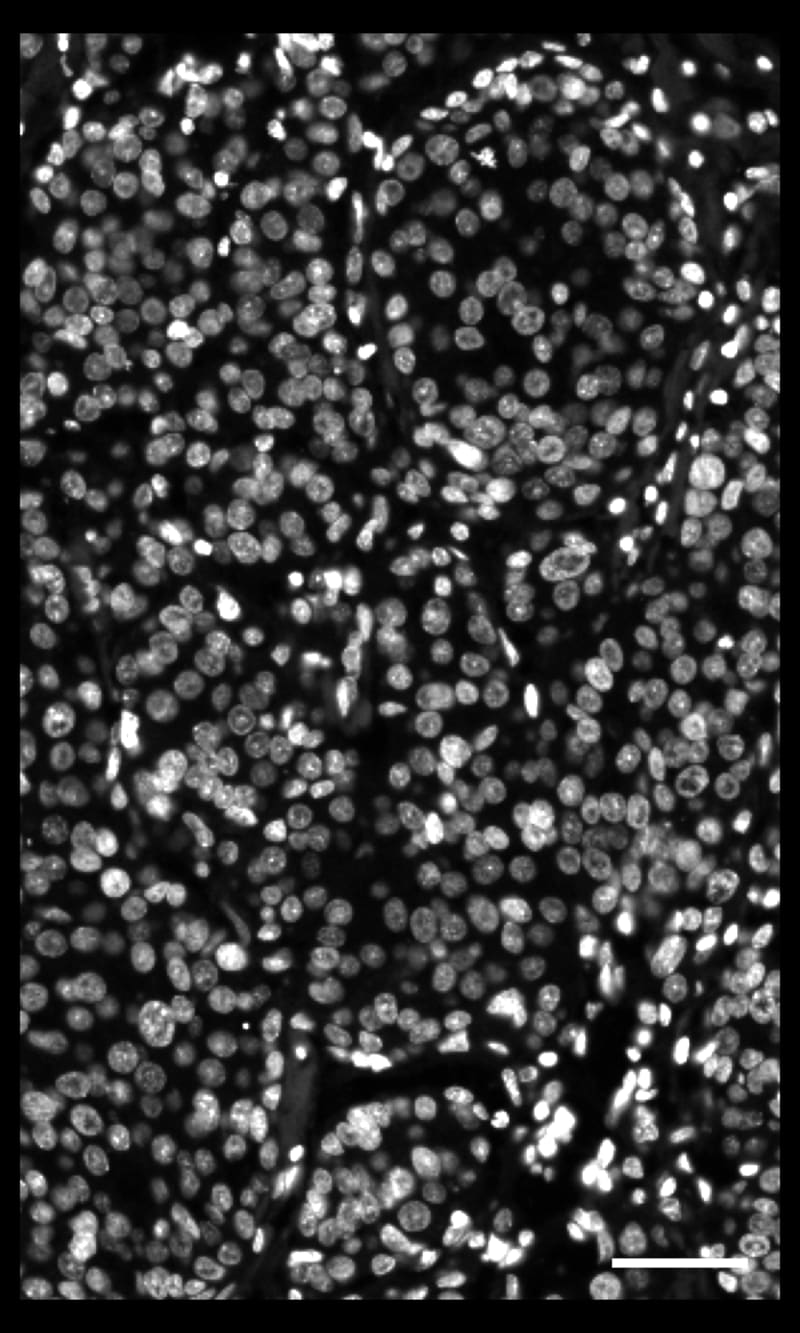
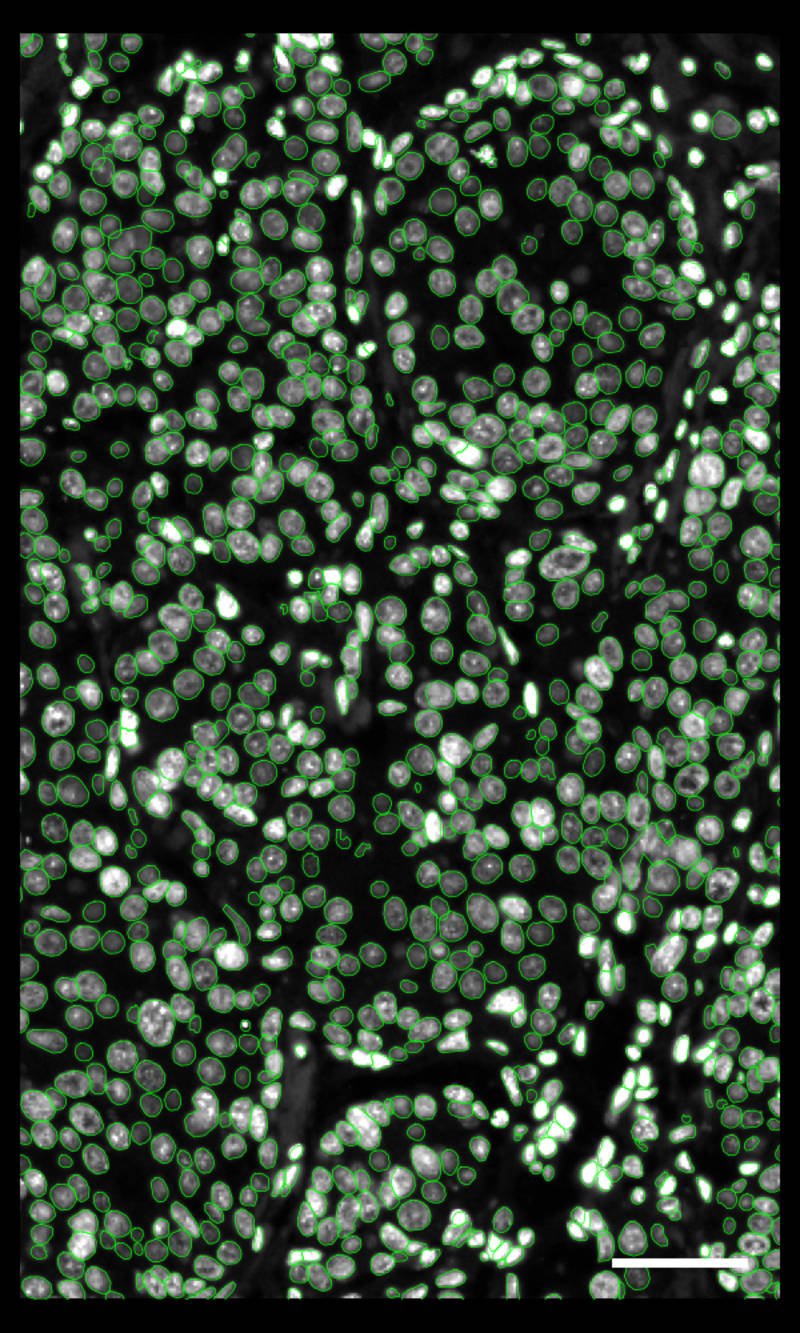
DAPIベースの細胞セグメンテーション
Xeniumでは、社内トレーニングモデルを使用してDAPI画像で核を区分します。Xenium解析ソフトウェアにより、通常とは違う形の核の区分も正確に行えます。核からの拡大された細胞エリアに対して転写産物のマッピングを行います。
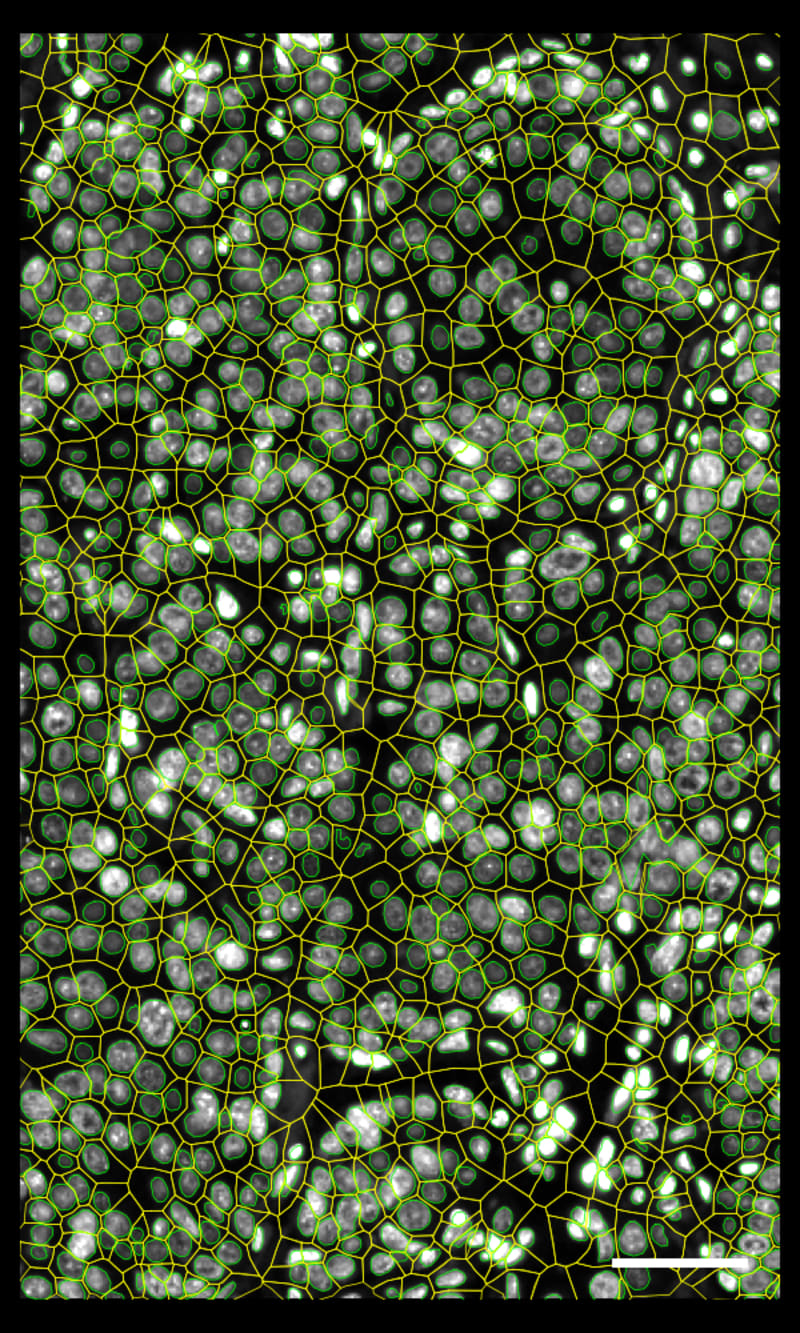
Xeniumの出力を一般に使用されているシングルセルデータ解析ソフトウェアで解析し、クラスターのアノテーションや発現変動遺伝子の同定を行えます。
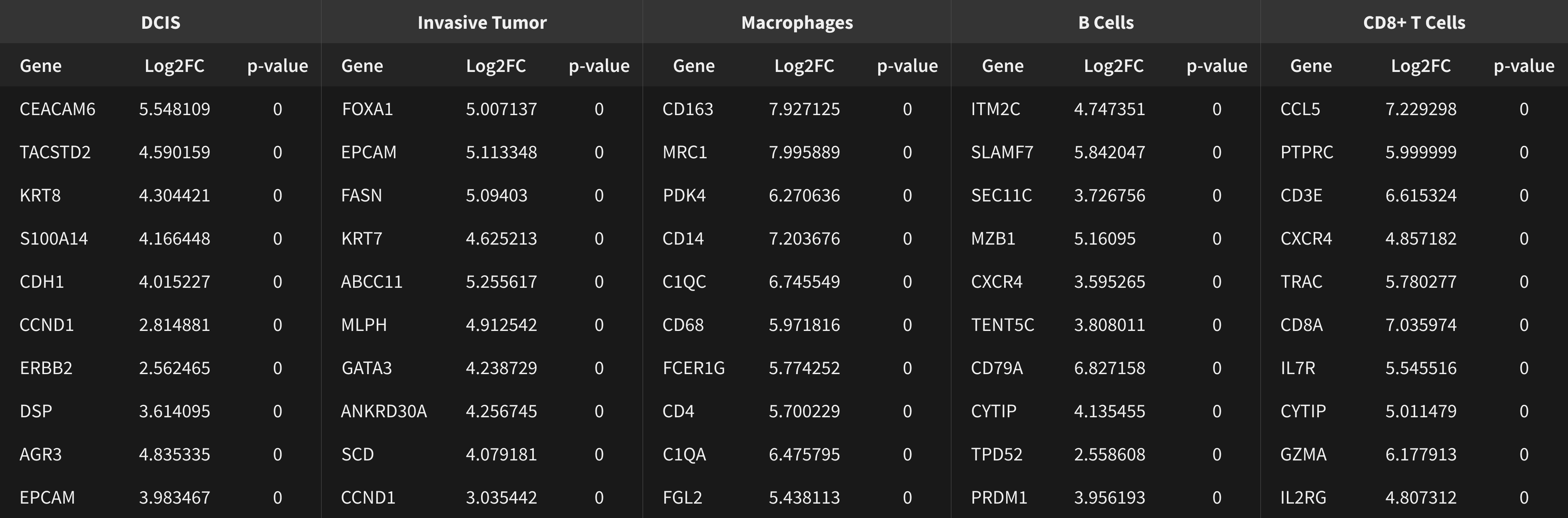
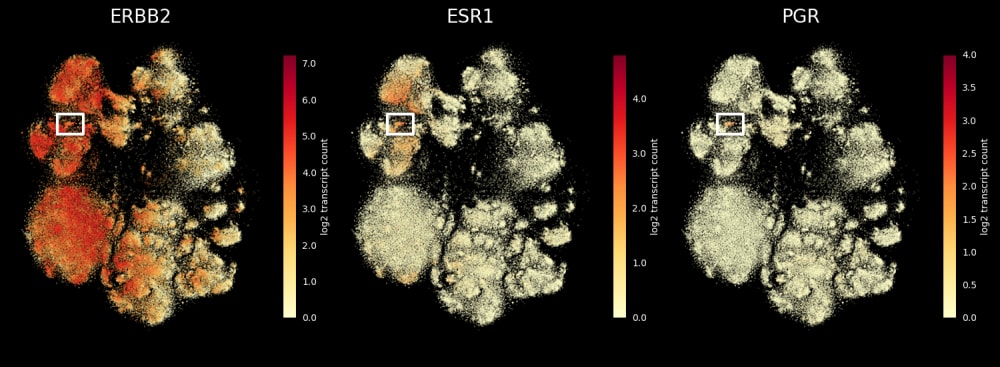
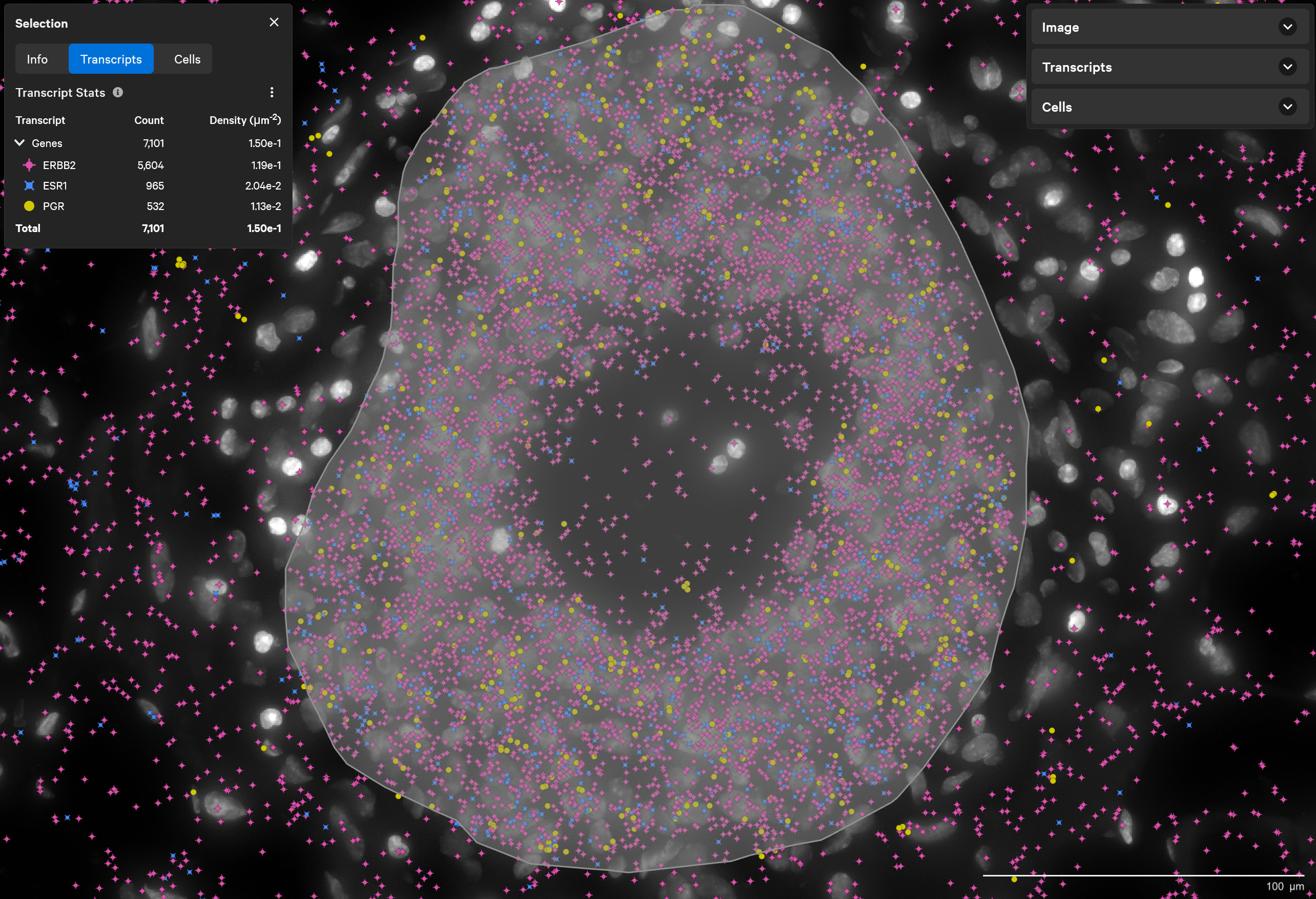
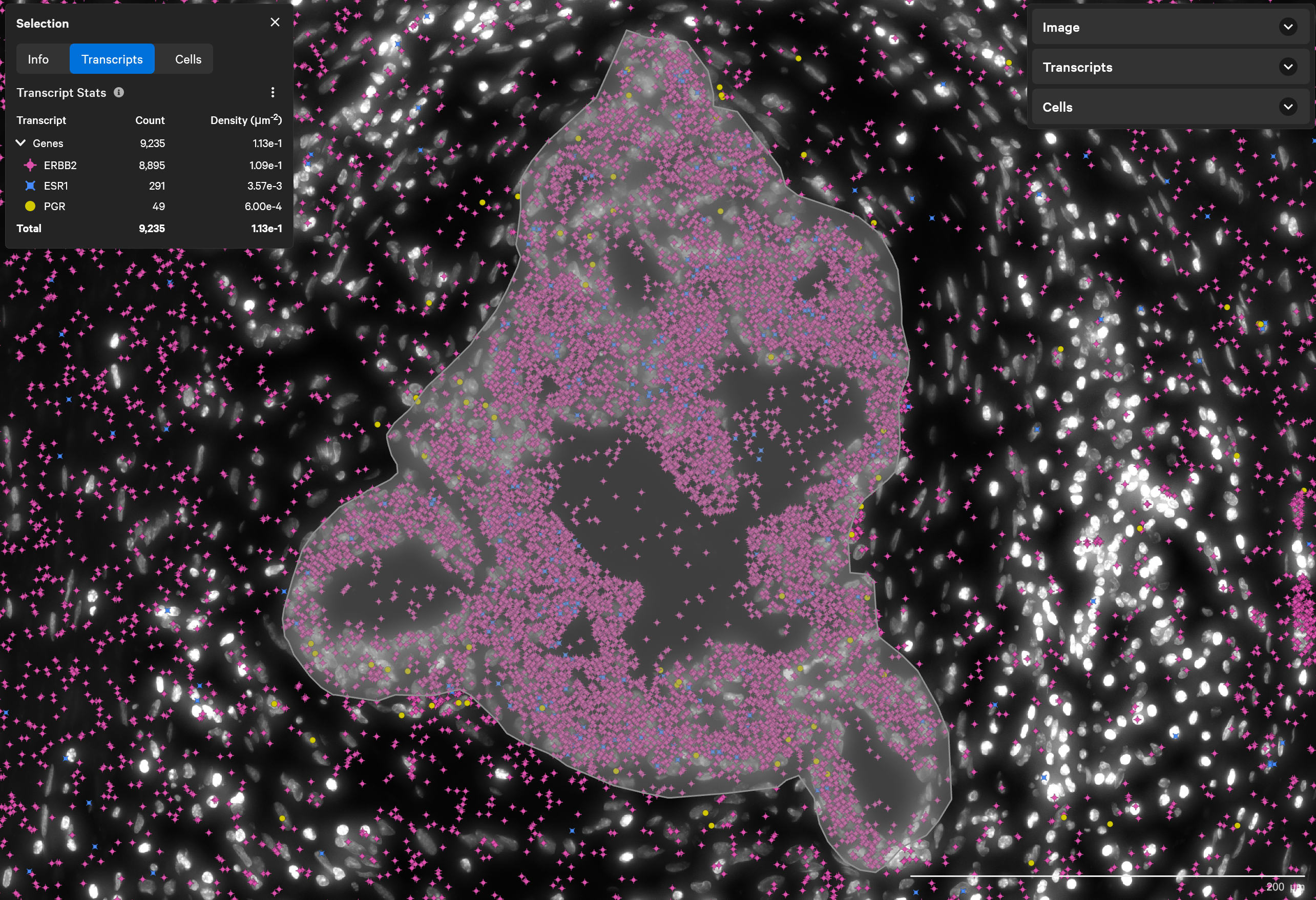
稀なトリプル陽性細胞の発見
ERBB2(HER2)陽性、 ESR1(ER)陽性、PGR(PR)陰性乳腺腫瘍に分類されるものに稀に含まれるトリプル陽性細胞(3種類のホルモン受容体のすべてを発現している細胞)を、Xeniumで発見します。