Single cell and spatial analysis empower new potential modes of treatment for autoimmune disease
Guest author: jamie-schwendinger
Explore the perplexing story of autoimmune disease, and how single cell and spatial analysis of conditions like rheumatoid arthritis, multiple sclerosis, type 1 diabetes, lupus, and Crohn’s disease are giving researchers new insights into treatment. We highlight the work our customers have done to understand autoimmune disorders, in honor of National Autoimmune Diseases Awareness Month.
Autoimmune diseases may represent the worst type of betrayal. The body’s own defense system, presumed to protect from harm and kill invaders, instead turns on its host. This internal malfunction manifests in a number of different ways, with diseases localized to specific organs or broadly systemic, and through varying mechanisms. At their core, however, autoimmune diseases arise from a defect in immune tolerance.
Immune tolerance is the process by which the body prunes the immune system to keep it from attacking self tissues. Central tolerance operates during immune cell development. In the thymus, T cells—a type of white blood cell that rouse targeted attacks on faulty host cells, like cancer cells or cells infected by a virus—that express cell surface receptors activated by ‘self’ antigens are destroyed or reprogrammed into T regulatory cells. B cells produced in the bone marrow face a similar fate, where cells expressing self-recognizing antibodies either undergo apoptosis or receptor editing. However, central tolerance is imperfect: like a leaky faucet aberrantly releasing droplets of water, self-recognizing immune cells can escape the defensive barriers of central tolerance. In fact, one study of healthy adults showed that the prevalence of self-recognizing CD8+ T cells was comparable to cells that recognized novel viral particles (1).
How do we keep all these escaped, self-reactive cells from mounting an immune response? One way is through a secondary gatekeeper: peripheral tolerance. Mechanisms of peripheral tolerance include inhibitory molecules, anergy, ignorance, and suppression by regulatory T cells. Inhibitory molecules expressed on the surface of T and B cells include CTLA-4, PD-1, and VISTA, all of which suppress immune response. Anergy is “an acquired state of functional unresponsiveness” and impacts both T and B cells. Ignorance is a lack of inflammatory response to antigens by circulating immune cells, and includes physical sequestration of antigens in areas like the central nervous system, testis, and eye. Regulatory T (T reg) cells suppress immune response by inhibiting T-cell proliferation and cytokine production. They are characterized by expression of forkhead box P3 (FOXP3), a transcription factor essential for T reg production and function (1).
However, even with these checks in place, tolerance can go awry.
Over 80 inflammatory disorders have been identified, and together they make up autoimmune disease. These disorders arise from defects in tolerance and are characterized by inappropriate inflammation. Affecting 7–9% of the population, though more prominently in women (1), autoimmune diseases result from a combination of genetic and environmental factors, lack a clear cause, and have no cures.
In the following studies, we explore several autoimmune diseases, including those that affect specific organs—Crohn’s disease, Type 1 diabetes, and multiple sclerosis—and others that spread to multiple tissues in the body—lupus and rheumatoid arthritis. Now, with the help of single cell and spatial analysis, researchers can interrogate and master the underlying biology of these perplexing disorders. And as these studies demonstrate, researchers are empowered to uncover new possible routes of treatment, offering hope for alternative treatment options beyond managing the symptoms of autoimmune disease.
Redefining treatment for organ-specific autoimmune disease
Irritable bowel disease (IBD) affects 3 million people in the US, and includes Crohn’s disease (impacting over half a million people) and ulcerative colitis. Crohn’s disease is characterized by inflammation of the intestinal wall and can cause a variety of issues, from diarrhea and abdominal pain, to complications like bowel obstruction, intestinal abscesses or ulcers, and malnutrition. There are no cures for Crohn’s disease, but current treatment options include anti-inflammatory medicines, and biologics like anti-TNF. Biologics can help patients go into remission, and provide relief to those who failed to respond to other treatments. Despite this treatment progress, however, 20–30% of ileal Crohn’s disease patients never respond to anti-TNF (2).
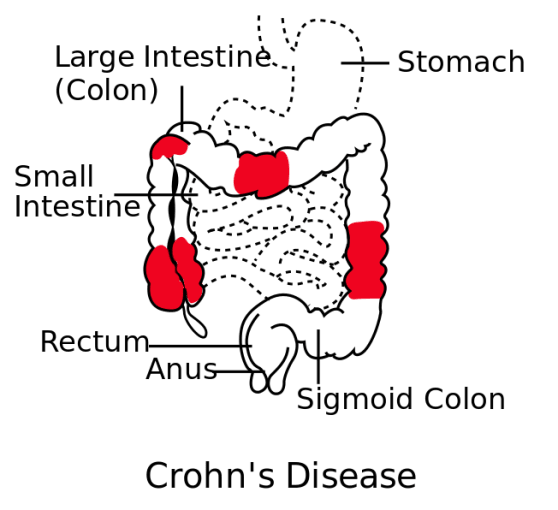
To understand why anti-TNF therapy is successful in some cases, but provides no relief in others, Jerome Martin and colleagues at the Icahn School of Medicine at Mount Sinai (New York, NY, USA) used single cell sequencing to characterize the cellular environment of inflamed and normal intestines from ileal Crohn’s disease patients. Combining single cell analysis of 11 patients with bulk sequencing and clinical evaluation from a larger (n=199) cohort, they identified a cellular signature unique to some patients. This signature included inflammatory macrophages, IgG plasma cells, and highly activated T cells. Although the presence of this cell module did not correlate with disease severity, it did appear prognostic of patients who would fail to respond to anti-TNF therapy. With the ability to predict patient response, clinicians may have the information they need to tailor therapeutic plans, avoid certain drug trials, or propose novel combination anti-TNF therapies that address the underlying cellular and molecular basis for resistance to anti-TNF blockade (2).
Another organ-specific autoimmune disease, type 1 diabetes is caused by the inability to produce insulin. The pancreas contains distinct structures called pancreatic islets. These islets are made up of different types of cells that produce critical hormones, including glucagon-producing alphas cells and insulin-producing beta cells. During type 1 diabetes, the immune system attacks and nullifies pancreatic beta cells, destroying the body’s insulin source. Current treatment for type 1 diabetes includes taking insulin and strict dietary monitoring, but the exact causes and mechanisms of the disorder are still poorly understood. Like Crohn’s disease, there is no cure.
For National Diabetes Month last November, the 10x Genomics blog highlighted two stories of researchers discovering novel approaches to potentially treat diabetes, each enabled by single cell sequencing. In the first study, researchers successfully reprogrammed human pancreatic alpha cells to sense glucose and make insulin—a job normally performed by beta cells. Single cell gene expression profiling helped the team identify several genes that may be responsible for this transition. These genes now represent possible target pathways for pharmacological intervention or gene therapy in the treatment of type 1 diabetes (3).
A second group of researchers also used single cell gene expression profiling to identify a previously uncharacterized beta-cell subpopulation with inflammation-triggering senescent signatures. Their finding suggested that senescent cells may drive autoimmunity in type 1 diabetes. If this is the case, then clearing these cells—in essence, supplementing a normal function of the immune system that has gone awry in the disease—could represent an alternative target of clinical intervention (4).
The brain is also susceptible to autoimmune disease. Multiple sclerosis (MS) is caused by an immune-mediated attack on the myelin sheath of neurons in the central nervous system. This aberrant immune response specifically targets oligodendrocytes, myelin-generating cells, and can lead to scattered scarring throughout the brain. Symptoms of MS arise from degradation in signal transmission as a result of neuronal damage and include fatigue, dizziness, pain or itching, and cognitive changes such as difficulty learning and remembering new information. A recent study estimates the prevalence of multiple sclerosis in 2017 in the United States at almost one million people, making the need for better treatment options even more pressing (5).
While current treatment options focus on managing symptoms, slowing progression, and reducing inflammation, recent studies have helped to characterize the underlying cellular heterogeneity and molecular dysregulation within MS lesions, and may guide more effective therapeutic intervention. As described in a blog post last year, researchers at UCSF performed single nucleus RNA-sequencing on frozen human brain samples, and found that neurons are differentially susceptible to multiple sclerosis lesions. Single nucleus analysis revealed that upper cortical neurons upregulated genes for self-antigen presentation to immune cells, indicating their particular vulnerability to degeneration and inflammation. Moreover, the researchers identified a subset of reactive microglial cells located on the edge of MS lesions in the subcortex that expressed genes associated with phagocytosis and carried ingested myelin transcripts in their nuclei. These findings clarify some of the transcriptomic changes and cellular dynamics that drive the progression of MS, and suggest that inflammation-prone cortical neuron populations could benefit from therapies that deplete local immune cells (6).
Fighting back against systemic autoimmune disease
Autoimmune diseases can also be systemic, impacting tissues and organs throughout the body. In systemic lupus erythematosus (lupus), autoantibodies provoke an inflammatory immune response in multiple organs, potentially affecting the skin, lungs, brain, joints, blood vessels, and kidney.
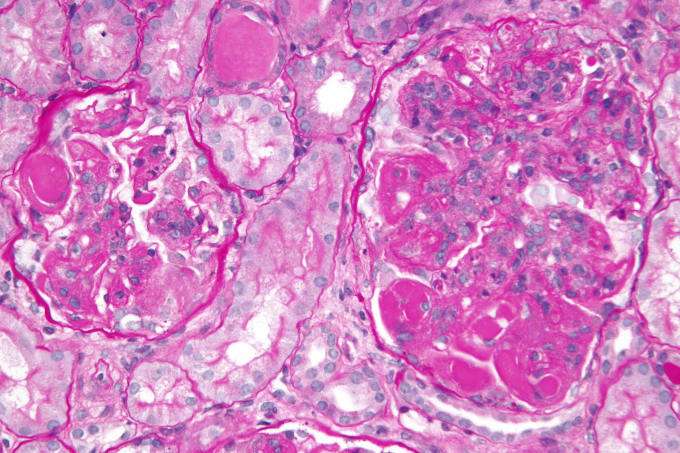
One of the more common complications of systemic lupus erythematosus is lupus nephritis: kidney malfunction or failure caused by autoimmune-induced inflammation of the kidneys. Symptoms include bloody or foamy urine, high blood pressure, and swelling of the hands, ankles, and feet. As with other autoimmune diseases, there is no cure for lupus nephritis, and treatment focuses on dampening immune responses and addressing the symptoms, with varying efficacy.
Single cell analysis, however, continues to open doors for the discovery and development of novel treatments for lupus nephritis. Using single cell transcriptional profiling, researchers from the Broad Institute of MIT and Harvard were able to reveal the complexity of immune populations in diseased kidneys. They identified 21 subsets of leukocytes active in disease, and noted heightened expression of chemokine receptors CXCR4 and CX3CR1 on immune cells. Corresponding expression patterns of the CXCR4 ligand showed that it derived mainly from epithelial cells and M2-like macrophages, suggesting these cell types coordinate traffic of immune cells infiltrating the kidney with a targetable chemokine signaling pathway. Explore an overview of this study in our Research Snapshot (7).
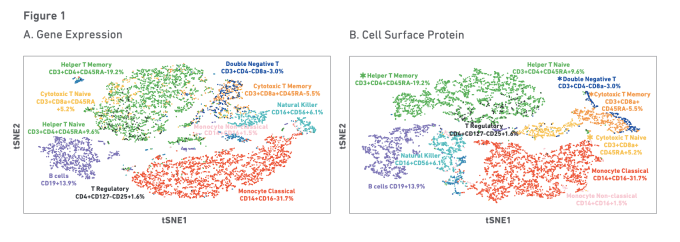
Researchers are also working to improve vaccine efficacy in patients with lupus. A recent study out of the NIH Center for Human Immunology focused on the underlying cellular and transcriptional signatures predictive of a healthy individual’s antibody response to both yellow fever and influenza vaccinations. Using multiomic single cell profiling of gene expression and surface proteins on blood samples from healthy influenza vaccine responders, researchers identified immune signatures related to activation of a plasmacytoid dendritic cell–type I IFN–T/B lymphocyte network. Evaluation of individuals with clinically quiescent systemic lupus erythematosus (SLE) revealed the same immune signature. In fact, sustained activity of the cells in this network was shown to drive disease severity, suggesting regulation of these immune baseline states could mitigate or alleviate autoimmune disease activity (8).
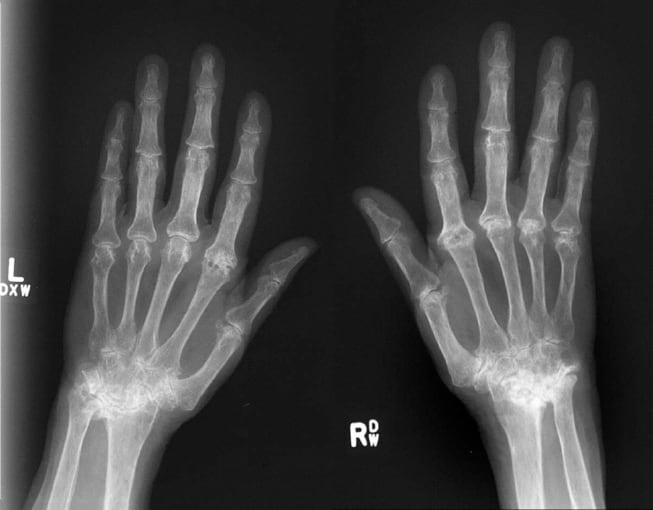
Another systemic autoimmune disease, rheumatoid arthritis is most commonly associated with inflammation of the joints. Joint inflammation is caused by an immune attack on the synovial membrane, a structure that lines the joints and produces synovial fluid. Prolonged inflammation thickens the synovium, leading to cartilage destruction and bone erosion.
Normally, cartilage is avascular, making it a site of immune privilege. However, in rheumatoid arthritis, this barrier is broken, and immune cells infiltrate the joint. Two recent publications have addressed the heterogeneous composition of synovial joints, including the immune signatures of the disease, in complementary ways. The first used single cell sequencing to identify the different cellular players, and the second used spatial transcriptomics, now available as the Visium Spatial Gene Expression Solution from 10x Genomics, to take a picture of gene expression within the joint.
Stephan Culemann and fellow researchers in Erlangen, Germany, discovered that macrophages play two roles in rheumatoid arthritis. Using single cell sequencing with fluorescent microscopy, they examined the role of macrophages in articular joints after onset of arthritis, observing an influx of pro-inflammatory macrophages in the joint. However, they also identified a surprising protective role for macrophages. In the absence of inflammation, a distinct population of macrophages lined the synovial cavity as a membrane-like structure. These macrophages expressed tight junctions, forming a protective immunological barrier that physically secluded the joint and restricted the inflammatory reaction (9). With this new understanding of the functional diversity among macrophages, researchers may have the baseline knowledge necessary to begin exploring how macrophages could support treatment of arthritis, or identify other cases in health and disease in which tissue-resident macrophages modulate inflammation.
In a second study, Konstantin Carlberg and scientists at the Science for Life Laboratory and Karolinska Institutet in Stockholm, Sweden, identified distinguishing immune and cellular characteristics of rheumatoid arthritis compared to spondyloarthritis, a type of arthritis that typically attacks the spine. They found that rheumatoid arthritis showed an adaptive immune response along with overrepresentation of central memory T cells, while spondyloarthritis resulted in more tissue repair responses and increase in effector memory T cells. Using spatial transcriptomics, the team annotated synovial biopsies alongside spatial analysis of whole transcriptome sequencing, and focused their genetic comparison on regions of the tissue showing immune infiltrate. This allowed them to refine the list of genes associated with the disease, clarifying variation across rheumatoid arthritis samples and further distinguishing rheumatoid arthritis and spondyloarthritis (10). As these findings demonstrate, the ability to map spatially resolved gene expression data onto high-resolution histological images may further guide our understanding of the diverse pathophysiological mechanisms—including where inflammation manifests in tissue, through what cell type, and by what gene pathways—underlying complex heterogeneous autoimmune diseases.
Single cell and spatial technologies hold the promise to change how we diagnose and treat autoimmune diseases. With the robust information gleaned from the application of these tools, clinicians may be more equipped to both understand and treat the causal mechanisms underlying autoimmune disorders, rather than chasing down the symptoms.
Explore more resources for immunology research →
References
- AN Theofilopoulos, DH Kono, R Baccala. The multiple pathways to autoimmunity. Nat Imm. 18, 716–724 (2017).
- C Martin et al., Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell. 178, 1493–1508.e20 (2019).
- K Furuyama et al., Diabetes relief in mice by glucose-sensing insulin-secreting human α-cells. Nature. 567, 7746 (2019).
- P Thompson et al., Targeted elimination of senescent beta cells prevents type 1 diabetes. Cell Metabolism. 29, 4 (2019).
- MT Wallin et al., The prevalence of MS in the United States: A population-based estimate using health-claims data. Neurology. 5, e1029–e1040 (2019).
- L Schirmer et al., Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature. 573 (2019).
- A Arazi et al., The immune cell landscape in kidneys of patients with lupus nephritis. Nat Imm. 20, 902–914 (2019).
- Y Kotliarov et al., Broad immune activation underlies shared set point signatures for vaccine responsiveness in healthy individuals and disease activity in patients with lupus. Nat Med. (2020).
- S Culemann et al., Locally renewing resident synovial macrophages provide a protective barrier for the joint. Nature. 572, 670–675 (2019).
- K Carlberg et al., Exploring inflammatory signatures in arthritic joint biopsies with Spatial Transciptomics. Sci Reports. 9, 18975 (2019).
