When single cell meets spatial, researchers map the crucial cellular processes in early human pregnancy
*Impact at a glance: Researchers from the Wellcome Sanger Institute and the University of Cambridge leveraged high-resolution single cell multiomics and spatial transcriptomics to understand the cellular relationships at the maternal–fetal interface driving successful implantation in the uterus and formation of the placenta.*
What happens when two powerful technologies join forces to resolve the “what” and the “where” of complex biology? We share those stories in our “When single cell meets spatial” series, highlighting studies that capture the best of both technologies—and how they can be even better together.
Mom and baby, getting close at early stages of pregnancy
The close connection between a mother and her child has profound significance, relationally and biologically. The development of the placenta is a collaborative project between the mother and fetus—both provide cells to form the temporary organ that protects and nurtures the developing fetus (1).
Specifically, trophoblast cells derived from the outer epithelial layer of the embryo kick off a major construction project following implantation into the mother’s uterus (2). Extravillous trophoblast (EVT) cells infiltrate the mucosal lining of the pregnant uterus, also called the decidua, and march towards a network of maternal arteries, which they subsequently transform into “high-conductance vessels” that will go on to supply oxygen and nutrients through the placenta to the fetus during pregnancy (2).
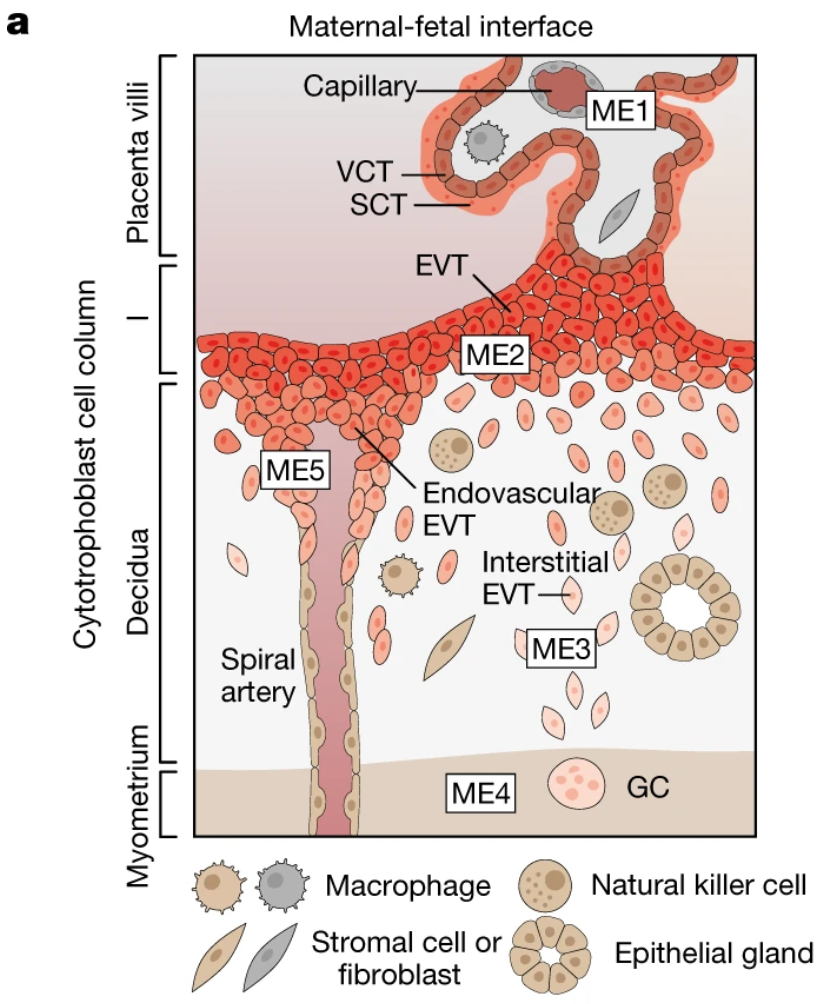
These steps to restructure maternal tissue and establish the placenta at the intimate meeting point between the mother and fetus are also implicated in common, and sometimes deadly, pregnancy disorders. This includes preeclampsia—a condition that causes irregular blood pressure in the mother, among other symptoms, likely as a result of improper remodeling of the maternal arteries during placentation, the formation of the placenta (3,4).
Deeply characterizing the cellular relationships at the maternal–fetal interface during placentation is crucially important not only to understand the mechanisms underlying pregnancy disorders, but also to identify interventions that can ensure healthier, safer pregnancies.
Single cell and spatial map how two worlds collide at the maternal–fetal interface
The maternal–fetal interface was the subject of a recent study for a team of researchers co-led by Drs. Anna Arutyunyan, Kenny Roberts, Kevin Troulé, Frederick Wong, and Megan Sheridan and corresponding author Dr. Roser Vento-Tormo, from the Wellcome Sanger Institute and the University of Cambridge (2). In order to better understand the interactions between fetal trophoblast cells and maternal decidual cells during placentation, the team leveraged high-resolution single cell multiomics and spatial transcriptomics to study the implantation site in first-trimester pregnant hysterectomies (2).
Their analysis combined previous single cell data and/or newly sampled placental-decidual tissue collected from 18 total donors, at 5–13 post-conception weeks (PCW), including three archival tissue blocks collected before 2006. Though these archived samples had been frozen for well over a decade, the researchers were able to isolate high-quality single cell and nuclei data due to proper long-term storage practices and the sensitivity of the Chromium Single Cell Gene Expression and Single Cell Multiome ATAC + Gene Expression assays (2).
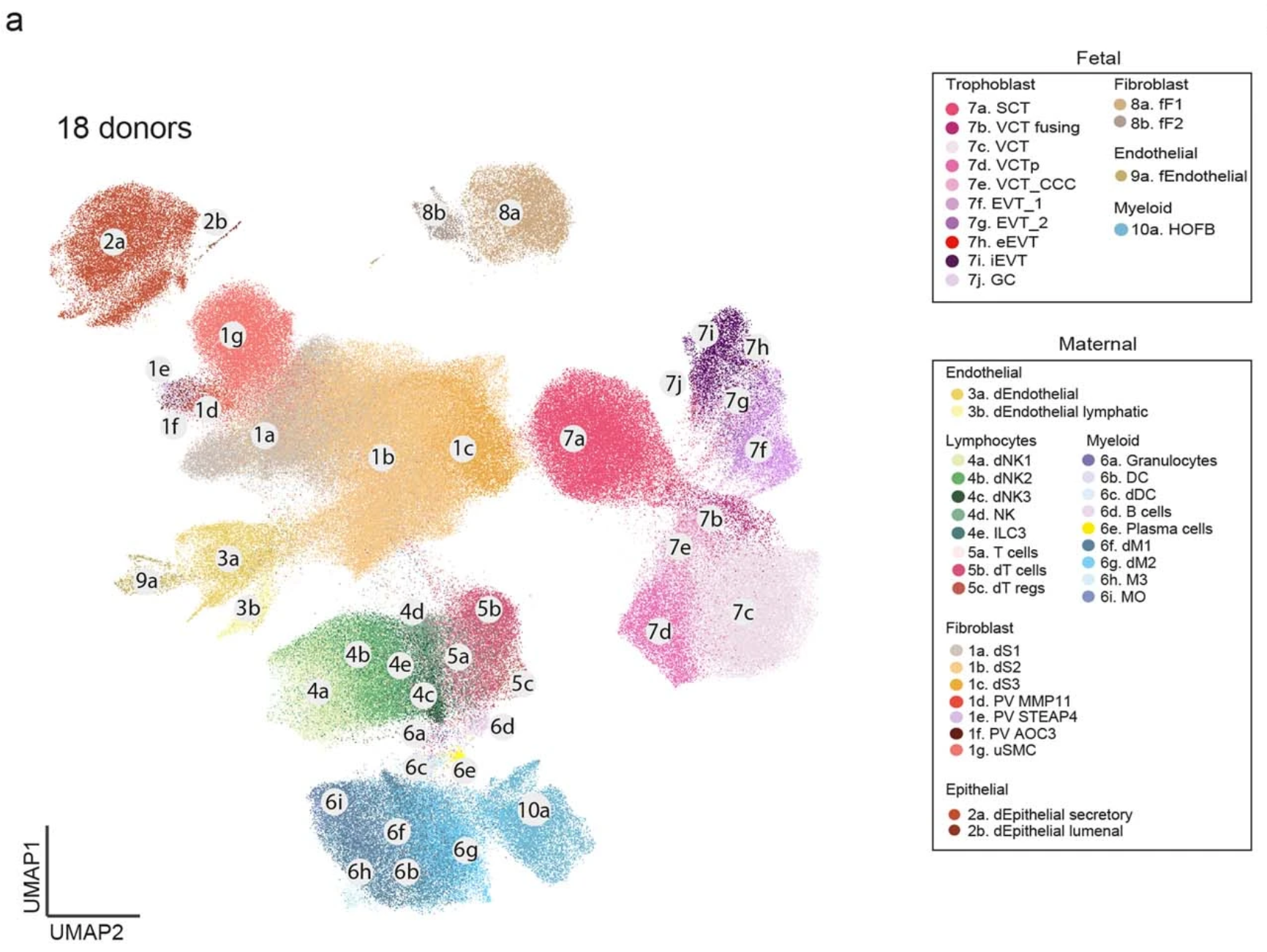
Using single cell gene expression data and analysis tools like CellPhoneDB v4, the team studied ligand–receptor interactions in the maternal–fetal interface that likely affect trophoblast invasion. They observed ligand–receptor interactions between CXCL16+CD14+ decidual macrophages and CXCR6+HLA-G+ EVT cells that served to upregulate placental genes in trophoblast cells, including genes responsible for increased motility (2).
They also placed tissue sections from a subset of the archival blocks on Visium slides for analysis by Visium Spatial Gene Expression, with paired morphological annotation of the fetal (placenta) and maternal (decidua and myometrium) tissue regions within the same slides, including four consecutive sections from donor P13 (2).
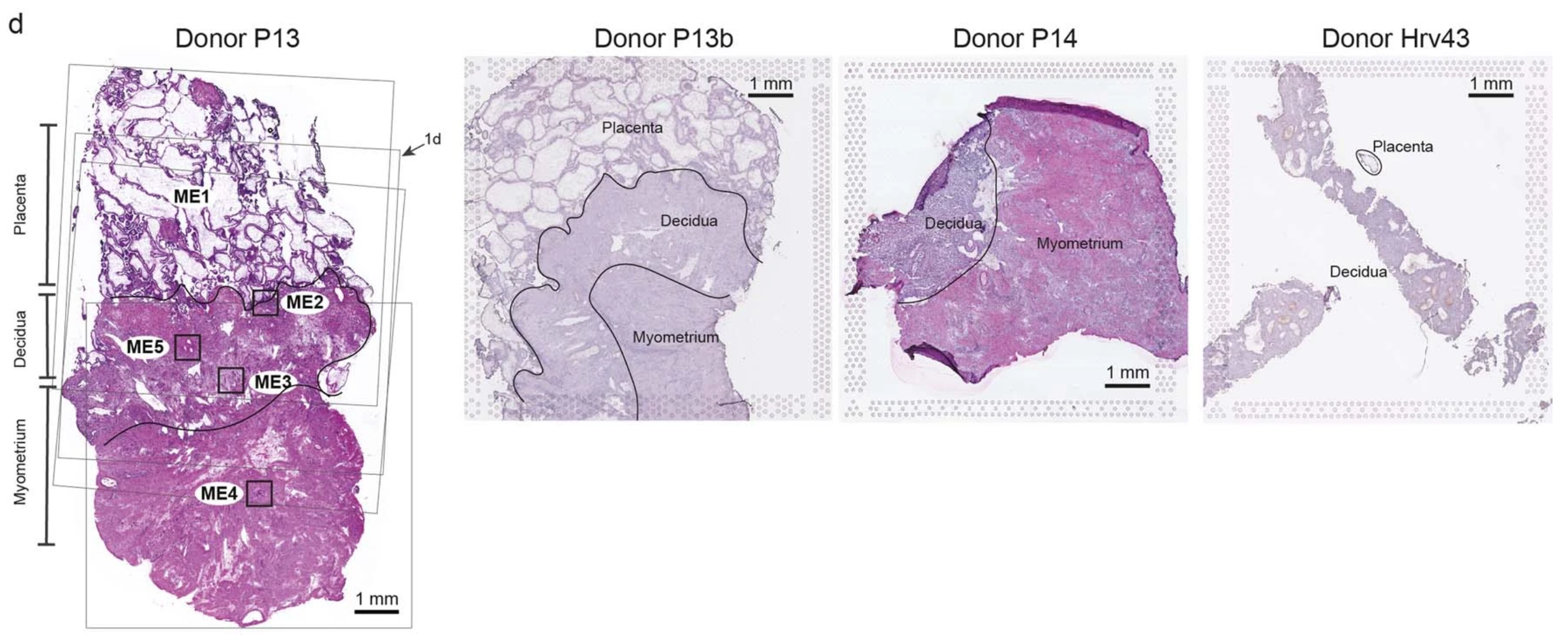
Using data analysis tools like cell2location, the team integrated single cell multiomic and spatial gene expression data to assess cell state densities within a given Visium spot. This analysis provided a rich spatial map of unique cell types and their gene expression signatures within the complex and changing architecture of the maternal–fetal interface.
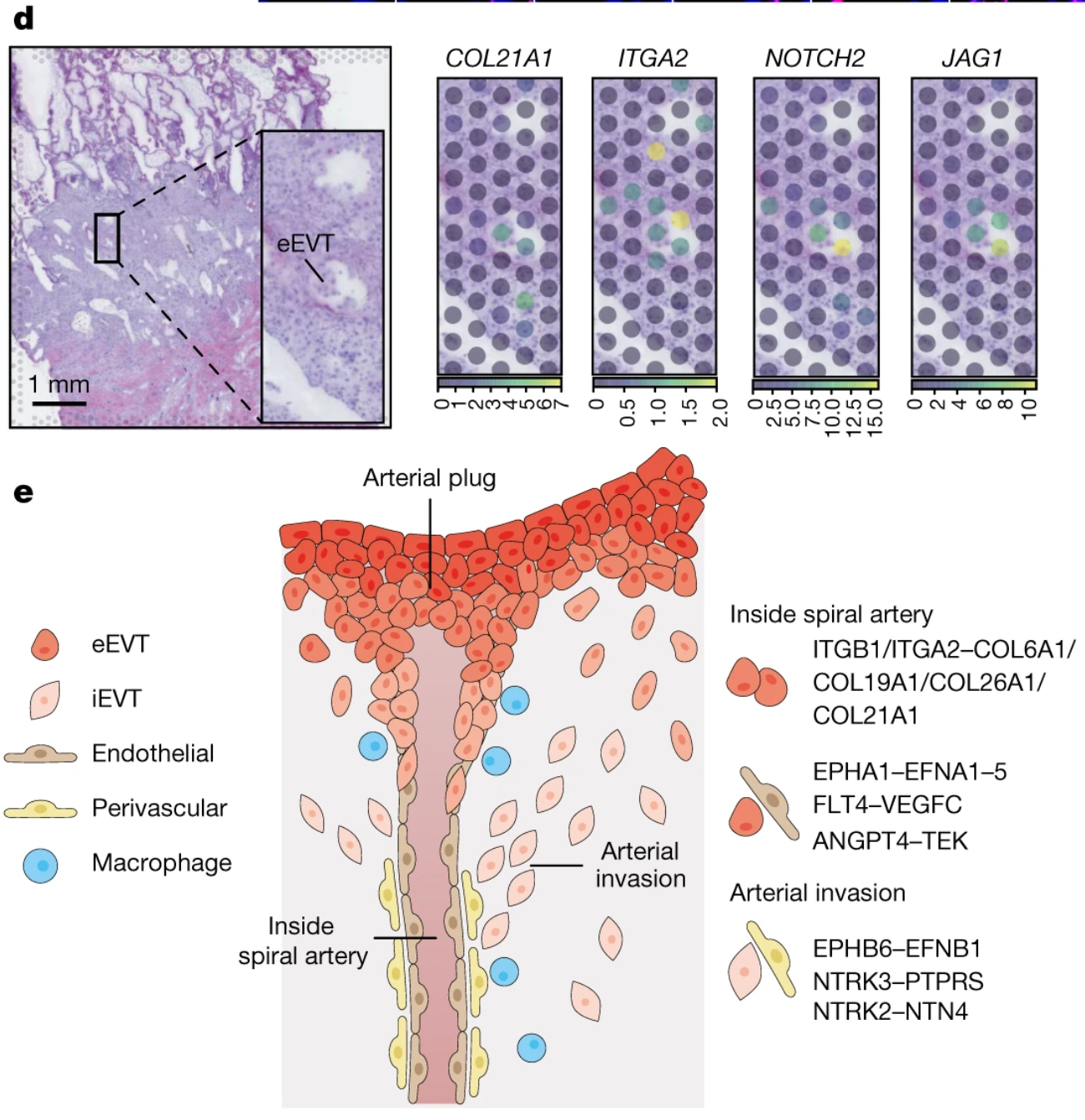
With the unbiased whole transcriptome data they derived from these assays, the team discovered a potential cellular and molecular mechanism for the transformation and plugging of the maternal spiral arteries after implantation. Zooming in on the spatial locations of trophoblast cells invading maternal spiral arteries, the team observed ligand–receptor interactions between interstitial EVT cells (iEVTs) expressing EFNB1 and maternal perivascular cells expressing the cognate receptor gene, EPHB6, which could drive iEVT tropism towards the arteries (2). The team noted other complementary ligand–receptor interactions between endovascular EVT cells (eEVT), which form a plug in spiral arteries during the reconstruction process, and endothelial cells that compose maternal vasculature (2).
Moreover, the team observed extracellular matrix component interactions, including COL21A1–ITGA2, and active Notch signaling, mediated by JAG1 and JAG2 ligand genes and NOTCH2 and NOTCH3 receptor genes, at the spatial location of arterial invasion. Notch signaling is a known mediator of extracellular matrix remodeling that occurs during angiogenesis—the formation of new blood vessels—suggesting these are both likely important signaling pathways mediating maternal arterial transformation (2,5).
Just the beginning of what we know about human pregnancy and delivery
“There has been a delay in our basic understanding of reproductive tissues. Now with the application of these novel technologies we can get more insight...”
“...which will have a huge impact on women’s health,” said Dr. Roser Vento-Tormo, Group Leader of Cellular Genetics at the Wellcome Sanger Institute and corresponding author for this study, in an interview with 10x Genomics.
There is still a lot to learn about many areas of women’s health, including pregnancy. But scientists are making great strides, with the application of high-resolution single cell and spatial tools, by mapping the specific cell types, cellular relationships, and signaling pathways that underlie successful human reproduction.
These insights come at the early stages of pregnancy, as in this study, but also at the end stages of pregnancy, with a recent Clinical and Translational Medicine paper employing single cell and spatial transcriptomics to characterize the immune landscape of myometrium tissue during labor, including the inflammatory processes that help to induce labor (6).
We celebrate the research these teams have contributed to the goal of advancing not only women’s health, but also the health and progress of society as a whole.
See how single cell and spatial technologies are helping us open up the black box of human embryonic development. Continue reading with this next blog.
Interested in learning more about the value of adding spatial context to single cell data? Explore the products discussed in this blog:
- Chromium Single Cell Gene Expression
- Chromium Single Cell Multiome ATAC + Gene Expression
- Visium Spatial Gene Expression
Download the Visium Essentials Guide to explore our direct-placement and Visium CytAssist–enabled spatial transcriptomics assays, how they work, and what tissues and species they’re compatible with.
References:
- Levine N. Five things we know about the placenta—and a few we wish we did. Cedars Sinai Discoveries Magazine (Feb 12, 2021).
- Arutyunyan A, et al. Spatial multiomics map of trophoblast development in early pregnancy. Nature 616: 143–151 (2023). doi: 10.1038/s41586-023-05869-0
- Brosens I, et al. The “great obstetrical syndromes” are associated with disorders of deep placentation. Am J Obstet Gynecol 204: 193–201 (2011). doi: 10.1016/j.ajog.2010.08.009
- https://www.mayoclinic.org/diseases-conditions/preeclampsia/symptoms-causes/syc-20355745
- Kretschmer M, et al. Matrix stiffness regulates Notch signaling activity in endothelial cells. J Cell Sci 136: jcs260442 (2023). doi: 10.1242/jcs.260442
- Ji K, et al. Integrating single-cell RNA sequencing with spatial transcriptomics reveals an immune landscape of human myometrium during labour. Clin Transl Med 13: e1234 (2023). doi: 10.1002/ctm2.1234
