On this page, you will learn the types and formats of data that you can import and export into and out of Loupe Browser.
In Loupe Cell Browser v3.1.0 and later, you can import custom projections. This allows you to compute a projection of the data using third-party software packages like Seurat or Scanpy to compute UMAP, t-SNE, PCA, or MDS projections. You can also import layouts from trajectory inference methods.
The CSV format for importing projections is as follows:
- First row: must be a header
- First column: barcodes that are at least a subset of the barcodes in the
.cloupefile - Second column: numbers which represent the X coordinates
- Third column: numbers that represent the Y coordinates
Here are the first few lines of an example UMAP projection:
Barcode X Coordinate Y Coordinate
AAACAAGCAAACTTCCACTTTAGG-1 5.603886971511171 -6.479924915
AAACAAGCAACATGATACTTTAGG-1 4.139326438982579 0.7246219486826851
AAACAAGCAATAGAGAACTTTAGG-1 5.408522227655841 1.7988092914659948
AAACAAGCACCTAAGAACTTTAGG-1 0.7652328791662211 -3.524468735
AAACAAGCAGAATGAAACTTTAGG-1 -0.749978434 -7.225456312
AAACAAGCATAGGGTCACTTTAGG-1 3.9925249310687185 0.6573977286625061
Download the full UMAP projection and import it for the lung carcinoma tutorial dataset in Loupe Browser. You can import new projections by clicking on the three dots to the right of the projection selector:
Importing the Custom-UMAP-Projection.csv should display this UMAP:
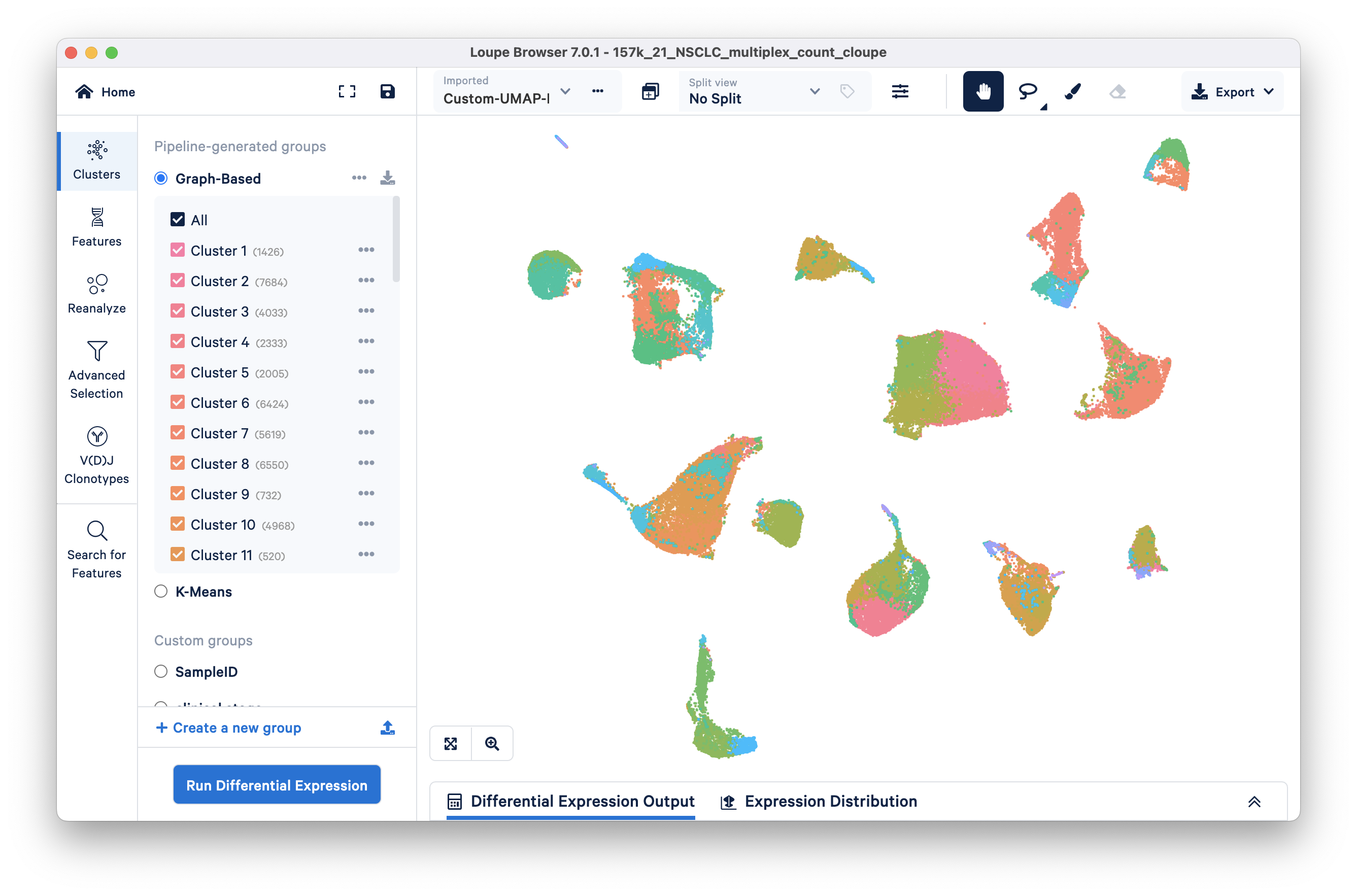
Once a custom projection is uploaded, you will also be able to rename it or delete it by clicking the same button that you used to import the projection.
Importing alternative projections is also described in the Navigation tutorial
In Loupe Browser v6.0 and later, you can also export the selected projection t-SNE or UMAP coordinates as a CSV file, including projections generated with the filtering and reclustering wizard, for further analysis in other software packages. Exporting projections is described in the Reanalyze tutorial.
Loupe Browser v4.0 and later allows you to integrate clustering data from other pipelines or scripts, or export manually assigned clusters into custom scripts. To do so, click on the action button to the right of the active cluster name, and select from one of the import/export options.
To create a clusters CSV file the format is as follows:
- The first row must be a header
- The first column header must be
barcodeand the column's contents must match at least a subset of the barcodes in the.cloupefile - Additional column headers are category names and the column's contents are cluster labels
Here is an example CSV:
Barcode,azimuth annotations
AAACAAGCAAACTTCCACTTTAGG-1,AT2
AAACAAGCAACATGATACTTTAGG-1,T cell lineage
AAACAAGCAATAGAGAACTTTAGG-1,T cell lineage
AAACAAGCACCTAAGAACTTTAGG-1,B cell lineage
AAACAAGCAGAATGAAACTTTAGG-1,Macrophages
AAACAAGCATAGGGTCACTTTAGG-1,T cell lineage
AAACAAGCATTCCTTTACTTTAGG-1,T cell lineage
The Azimuth cell type annotations CSV that was used in the lung carcinoma tutorial dataset is available for download.
There are two ways to create feature lists that you can use to quickly find cells of interest within your data, as described in the Identifying Cell Types portion of the tutorial. The first is to use the Gene/Feature search bar in Features Mode to search for individual features and build a list within Loupe Browser, whereas the second method involves creating a list of features (CSV) and importing it into Loupe Browser.
Both these methods are described in the Feature mode section of the Loupe Browser Navigation for Single Cell tutorial.
Filters can be imported, exported, and used across datasets similar to other features we have discussed on this page. However, we recommend that you do not manually create filters as they use a JSON specification that is specific to Loupe Browser and is subject to modification.