In this tutorial, you will learn how to run the spaceranger count pipeline on Visium Spatial Gene Expression data derived from a formalin fixed paraffin embedded (FFPE) mouse brain section.
To successfully run this tutorial, you should:
- Be comfortable in the Linux environment
- Have familiarity running command line tools
- Choose a compute platform
- Have access to a system that meets the minimum system requirements
Visium Spatial Gene Expression data from FFPE tissues are analyzed using the spaceranger count pipeline. The pipeline inputs a microscope image of the Visium slide (in TIFF or JPEG format), a reference, and FASTQ files, and performs alignment, tissue and fiducial detection, and barcode/UMI counting. Outputs include the feature-spot matrices, clustering and differential gene expression (DGE) which can be further analyzed and visualized in Loupe Browser.
In this tutorial, we will run the spaceranger count pipeline on a mouse brain FFPE section.
Key dataset features are:
- Tissue section of 5 µm thickness
- Section Orientation: Coronal
- H&E image
- Sequencing Depth: 28,826 reads per spot
- Sequencing Coverage: Read 1 - 28 bp (includes 16 bp Spatial Barcode, 12 bp UMI); Read 2 - 120 bp (transcript); i7 sample index - 10 bp; i5 sample index - 10 bp
- Visium Slide: V11J26-127
- Capture Area: B1
The following commands will be run in the working directory (spaceranger_tutorial) that was used to install spaceranger on a compatible compute platform.
Example FFPE dataset
Download with curl command:
# Create datasets folder
mkdir datasets
# Download FASTQ to datasets folder
curl https://cf.10xgenomics.com/samples/spatial-exp/1.3.0/Visium_FFPE_Mouse_Brain/Visium_FFPE_Mouse_Brain_fastqs.tar -o datasets/Visium_FFPE_Mouse_Brain_fastqs.tar
# Download image file to datasets folder
curl https://cf.10xgenomics.com/samples/spatial-exp/1.3.0/Visium_FFPE_Mouse_Brain/Visium_FFPE_Mouse_Brain_image.jpg -o datasets/Visium_FFPE_Mouse_Brain_image.jpg
# Expected output
% Total % Received % Xferd Average Speed Time Time Time Current
Dload Upload Total Spent Left Speed
5 4154M 5 218M 0 0 32.7M 0 0:02:06 0:00:06 0:02:00 33.0M
Alternatively, download with the wget command:
# Create datasets folder
mkdir datasets
# Download FASTQ to datasets folder
wget -P datasets/ https://cf.10xgenomics.com/samples/spatial-exp/1.3.0/Visium_FFPE_Mouse_Brain/Visium_FFPE_Mouse_Brain_fastqs.tar
# Download image file to datasets folder
wget -P datasets/ https://cf.10xgenomics.com/samples/spatial-exp/1.3.0/Visium_FFPE_Mouse_Brain/Visium_FFPE_Mouse_Brain_image.jpg
# Expected output
Resolving cf.10xgenomics.com (cf.10xgenomics.com)... 104.18.0.173, 104.18.1.173, 2606:4700::6812:1ad, ...
Connecting to cf.10xgenomics.com (cf.10xgenomics.com)|104.18.0.173|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 4356188160 (4.1G) [application/x-tar]
Saving to: ‘datasets/Visium_FFPE_Mouse_Brain_fastqs.tar’
37% [=======================> ] 1,649,129,803 207MB/s eta 13s
Reference data
Download the latest version of the mouse transcriptome reference from the Downloads page.
# Download mouse reference
curl -O https://cf.10xgenomics.com/supp/spatial-exp/refdata-gex-mm10-2020-A.tar.gz
# Expected output
% Total % Received % Xferd Average Speed Time Time Time Current
Dload Upload Total Spent Left Speed
1 9835M 1 158M 0 0 34.1M 0 0:04:48 0:00:04 0:04:44 34.1M
Probe set reference
Since this is a FFPE tissue sample, the assay uses a pair of oligonucleotide probes targeting protein-coding genes. In addition to the reference transcriptome, spaceranger count also requires the species-specific probe set reference file in CSV format. You can either download the probe set reference from the 10x support website or use the probe set references pre-bundled in Space Ranger.
# Method 1
# Download mouse probe set reference from support website
curl -O https://cf.10xgenomics.com/supp/spatial-exp/probeset/Visium_Mouse_Transcriptome_Probe_Set_v1.0_mm10-2020-A.csv
# Method 2
# Space Ranger 2.0 comes bundled with probe set files
## Source mouse probe set reference
~/spaceranger_tutorial/spaceranger-2.0.0/probe_sets/Visium_Mouse_Transcriptome_Probe_Set_v1.0_mm10-2020-A.csv
# Expected output
% Total % Received % Xferd Average Speed Time Time Time Current
Dload Upload Total Spent Left Speed
100 2257k 100 2257k 0 0 5579k 0 --:--:-- --:--:-- --:--:-- 5587k
In this tutorial, we will use the path associated with the first option.
Extract files
After downloading the required files, the contents of the tar files need to be extracted.
# Extract sample FASTQ files
tar -xvf datasets/Visium_FFPE_Mouse_Brain_fastqs.tar -C datasets/ && rm datasets/Visium_FFPE_Mouse_Brain_fastqs.tar
# Extract mouse reference transcriptome
tar -xzvf refdata-gex-mm10-2020-A.tar.gz && rm refdata-gex-mm10-2020-A.tar.gz
# Expected output
# Sample FASTQ files
Visium_FFPE_Mouse_Brain_fastqs/
Visium_FFPE_Mouse_Brain_fastqs/Visium_FFPE_Mouse_Brain_S3_L002_R1_001.fastq.gz
Visium_FFPE_Mouse_Brain_fastqs/Visium_FFPE_Mouse_Brain_S3_L001_I2_001.fastq.gz
Visium_FFPE_Mouse_Brain_fastqs/Visium_FFPE_Mouse_Brain_S3_L002_R2_001.fastq.gz
...
# Reference mouse transcriptome
refdata-gex-mm10-2020-A/
refdata-gex-mm10-2020-A/fasta/
refdata-gex-mm10-2020-A/fasta/genome.fa
...
Successful extraction will create two additional folders, highlighted in yellow, within the working directory.
1spaceranger_tutorial 2├── datasets 3│ ├── Visium_FFPE_Mouse_Brain 4│ └── Visium_FFPE_Mouse_Brain_image.jpg 5├── refdata-gex-mm10-2020-A 6├── Visium_Mouse_Transcriptome_Probe_Set_v1.0_mm10-2020-A.csv 7└── spaceranger-2.0.0
You can now build the spaceranger count command to run from your working directory (spaceranger_tutorial). If running from a different directory, amend the paths accordingly to avoid any errors.
spaceranger count --id="Visium_FFPE_Mouse_Brain" \
--transcriptome=refdata-gex-mm10-2020-A \
--probe-set=Visium_Mouse_Transcriptome_Probe_Set_v1.0_mm10-2020-A.csv \
--fastqs=datasets/Visium_FFPE_Mouse_Brain_fastqs \
--image=datasets/Visium_FFPE_Mouse_Brain_image.jpg \
--slide=V11J26-127 \
--area=B1 \
--reorient-images=true \
--localcores=16 \
--localmem=128
Below are brief descriptions of the command line options:
| Option | Description |
|---|---|
--id | The id must be a unique string and will be used to name the resulting folder with all of the pipeline outputs. |
--transcriptome | The path to the species-specific pre-compiled transcriptome files. You can provide the relative or absolute path to this folder. |
--probe-set | The absolute or relative path to the species-specific probe set reference file in CSV format. |
--fastqs | The path to the folder containing sample sequencing files in FASTQ format. |
--image | The path to a single brightfield image with H&E staining in either TIFF or JPEG formats. |
--slide | The Visium slide serial number. |
--area | The Capture Area identifier on the visium slide. It can be one of four values: A1, B1, C1 or D1. |
--reorient-images | Option to choose whether spaceranger should rotate and mirror the image to find the best fiducial alignment. Acceptable values are true or false with default being true. Useful to set to false when you are absolutely certain the fiducial corners in the input image are in the canonical positions (hourglass in top left corner) similar to included image Visium_FFPE_Mouse_Brain_image.jpg. Setting this to false will reduce pipeline runtime and prevent the pipeline from finding a fiducial alignment where the image is rotated/mirrored. |
--localcores | The number of CPU cores available to run the spaceranger count pipeline. |
--localmem | The max memory in GB available to run the spaceranger count pipeline. |
At the start of the run, you should see the preflight checks printed to the command line.
# Run spaceranger count
spaceranger count --id="Visium_FFPE_Mouse_Brain" \
--description="Adult Mouse Brain (FFPE) using Mouse WTA Probe Set" \
--transcriptome=refdata-gex-mm10-2020-A \
--probe-set=Visium_Mouse_Transcriptome_Probe_Set_v1.0_mm10-2020-A.csv \
--fastqs=datasets/Visium_FFPE_Mouse_Brain_fastqs \
--image=datasets/Visium_FFPE_Mouse_Brain_image.jpg \
--slide=V11J26-127 \
--area=B1 \
--reorient-images=true \
--localcores=16 \
--localmem=128
# Expected output
Martian Runtime - v4.0.5
Running preflight checks (please wait)...
Checking sample info...
Checking FASTQ folder...
Checking reference...
Checking reference_path...
Checking optional arguments...
...
Successful completion of the pipeline is indicated by a list of output files.
Outputs:
- Run summary HTML: /spaceranger_tutorial/Visium_FFPE_Mouse_Brain/outs/web_summary.html
- Outputs of spatial pipeline:
aligned_fiducials: /spaceranger_tutorial/Visium_FFPE_Mouse_Brain/outs/spatial/aligned_fiducials.jpg
detected_tissue_image: /spaceranger_tutorial/Visium_FFPE_Mouse_Brain/outs/spatial/detected_tissue_image.jpg
scalefactors_json: /spaceranger_tutorial/Visium_FFPE_Mouse_Brain/outs/spatial/scalefactors_json.json
tissue_hires_image: /spaceranger_tutorial/Visium_FFPE_Mouse_Brain/outs/spatial/tissue_hires_image.png
tissue_lowres_image: /spaceranger_tutorial/Visium_FFPE_Mouse_Brain/outs/spatial/tissue_lowres_image.png
cytassist_image: null
aligned_tissue_image: null
tissue_positions: /spaceranger_tutorial/Visium_FFPE_Mouse_Brain/outs/spatial/tissue_positions.csv
spatial_enrichment: /spaceranger_tutorial/Visium_FFPE_Mouse_Brain/outs/spatial/spatial_enrichment.csv
barcode_fluorescence_intensity: null
- Run summary CSV: /spaceranger_tutorial/Visium_FFPE_Mouse_Brain/outs/metrics_summary.csv
- Correlation values between isotypes and Antibody features: null
- BAM: /spaceranger_tutorial/Visium_FFPE_Mouse_Brain/outs/possorted_genome_bam.bam
- BAM BAI index: /spaceranger_tutorial/Visium_FFPE_Mouse_Brain/outs/possorted_genome_bam.bam.bai
- BAM CSI index: null
- Filtered feature-barcode matrices MEX: /spaceranger_tutorial/Visium_FFPE_Mouse_Brain/outs/filtered_feature_bc_matrix
- Filtered feature-barcode matrices HDF5: /spaceranger_tutorial/Visium_FFPE_Mouse_Brain/outs/filtered_feature_bc_matrix.h5
- Unfiltered feature-barcode matrices MEX: /spaceranger_tutorial/Visium_FFPE_Mouse_Brain/outs/raw_feature_bc_matrix
- Unfiltered feature-barcode matrices HDF5: /spaceranger_tutorial/Visium_FFPE_Mouse_Brain/outs/raw_feature_bc_matrix.h5
- Secondary analysis output CSV: /spaceranger_tutorial/Visium_FFPE_Mouse_Brain/outs/analysis
- Per-molecule read information: /spaceranger_tutorial/Visium_FFPE_Mouse_Brain/outs/molecule_info.h5
- Loupe Browser file: /spaceranger_tutorial/Visium_FFPE_Mouse_Brain/outs/cloupe.cloupe
- Feature Reference: null
- Target Panel file: null
- Probe Set file: /spaceranger_tutorial/Visium_FFPE_Mouse_Brain/outs/probe_set.csv
Waiting 6 seconds for UI to do final refresh.
Pipestance completed successfully!
After the run is completed, the working directory will have a new folder named V1_Adult_Mouse_Brain (value provided to --id argument) that contains all the metadata and outputs generated from the spaceranger count pipeline. We will highlight some key components of this folder:
Visium_FFPE_Mouse_Brain
├── _cmdline
├── _filelist
├── _finalstate
├── _invocation
├── _jobmode
├── _log
├── _mrosource
├── outs
├── _perf
├── _sitecheck
├── SPATIAL_RNA_COUNTER_CS
├── _tags
├── _timestamp
├── _uuid
├── Visium_FFPE_Mouse_Brain.mri.tgz
├── _vdrkill
└── _versions
Visium_FFPE_Mouse_Brain.mri.tgzcontains diagnostic information helpful to 10x Genomics support to resolve any errors_sitecheckcaptures the system configuration similar to sitecheck subcommand_timestampcontains information on pipeline runtimes. The runtime for the example dataset with the above configuration was 50:58_cmdlinecaptures thecountcommand provided to run the pipeline_versionscontains both thespacerangerand Martian versions used in the runoutscontains all the final pipeline generated outputs:
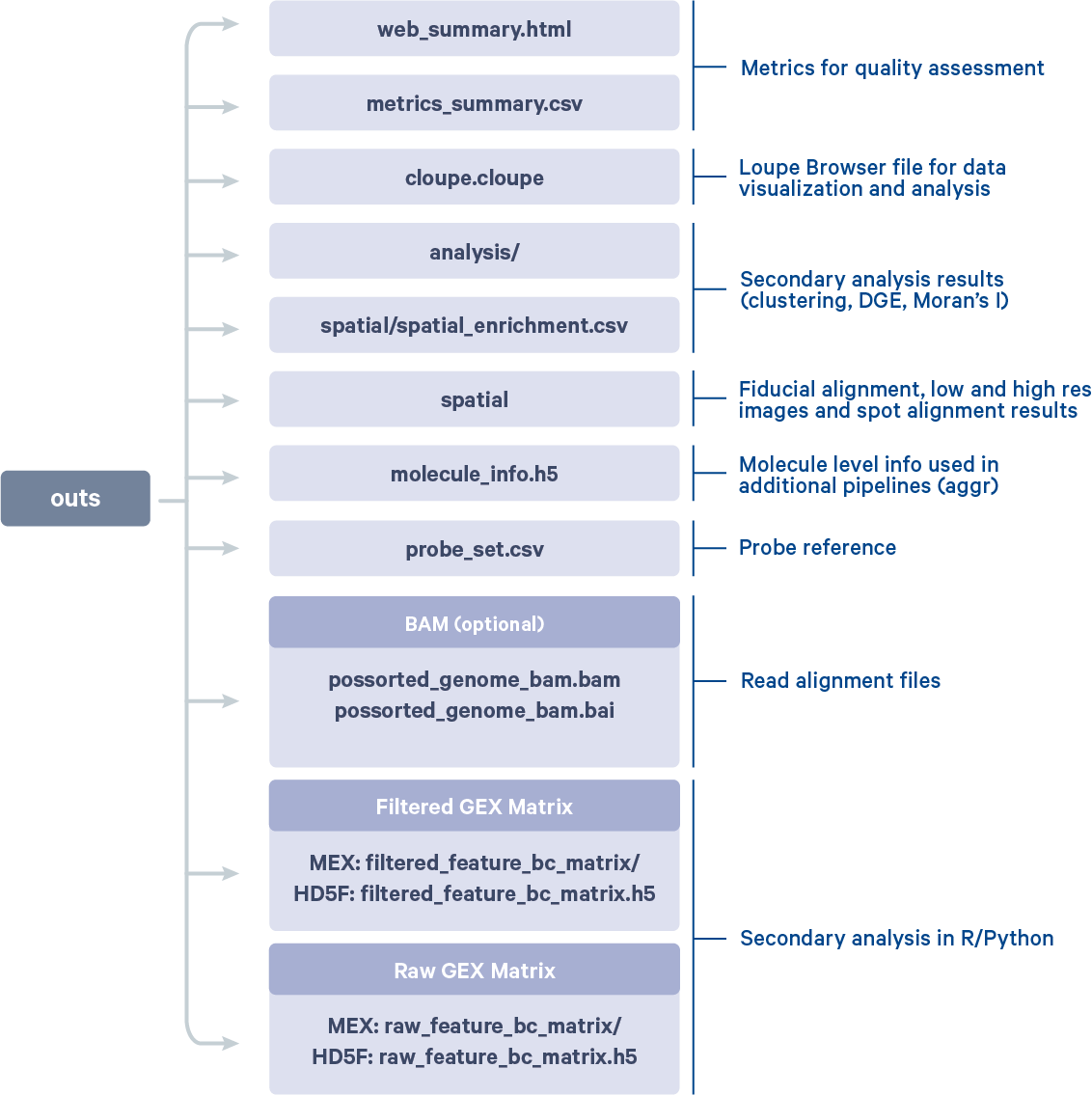
You can further explore and understand these results by
- Browsing the web_summary.html file.
- Opening the .cloupe file in [Loupe Browser](/support/software/loupe-browser/latest
- Referring to the Understanding Outputs page to explore individual files