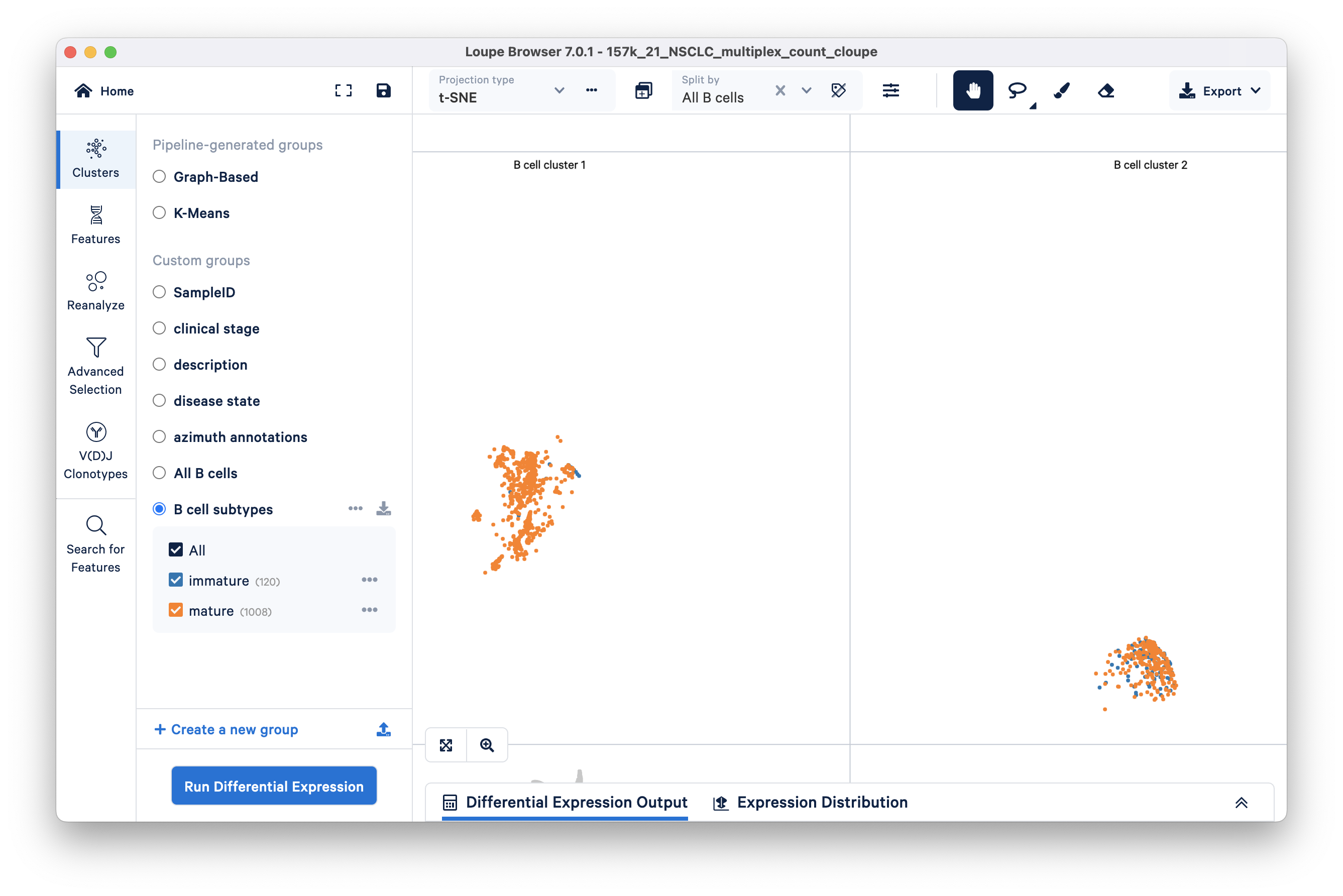
Learn how to find cell subtypes (immature and mature B cell) in the lung carcinoma tutorial dataset.
In the identifying cell types and exploring substructure section, we segmented B cells in t-SNE space by lassoing.
Next, we build on this work by finding and creating new clusters of immature and mature B cells within the previous B cell cluster we found. We will use the Advanced Selection mode and classify B cells as mature or immature depending on the presence (immature) or absence (mature) of the TCL1A gene.
Advanced Selection mode allows you to create boolean filters with one level of nesting. Each ruleset represents a grouping of rules which are joined by the same logical operator. The logical operator can be toggled between AND/OR.
When the first ruleset has more than one rule, you can add additional rulesets. To add rulesets, clicking the 'Add new ruleset' option located at the bottom of the panel. Rulesets operate independently from one another. Each ruleset may have its own logical operator.
Rulesets are all joined by the same logical operator. This can similarly be toggled between AND/OR using the dropdown at the top when there are greater than two rulesets. The options are:
- "All of the following" (AND) where a barcode is highlighted if all rulesets are true
- "Any of the following" (OR) where a barcode is highlighted if at least one ruleset is true
To filter for mature vs. immature B cells within B cell clusters 1 and 2, we will create two new filters containing two rulesets each.
Click Advanced Selection on the Mode Selector panel and rename the default Untitled Filter to Immature B cell subtypes.
Add the first cluster of interest (B cell cluster 1 created in the Explore Substructure tutorial) by clicking the blue 'Create new rule' button, then the 'IN [cluster name]' option, and finally enter the cluster name 'B cell cluster 1'.
To filter for immature B cells, click 'Create new rule' again and threshold barcodes by feature count > 0. After selecting this option, you will have to type in the TCL1A (feature name) in the Feature box as shown:
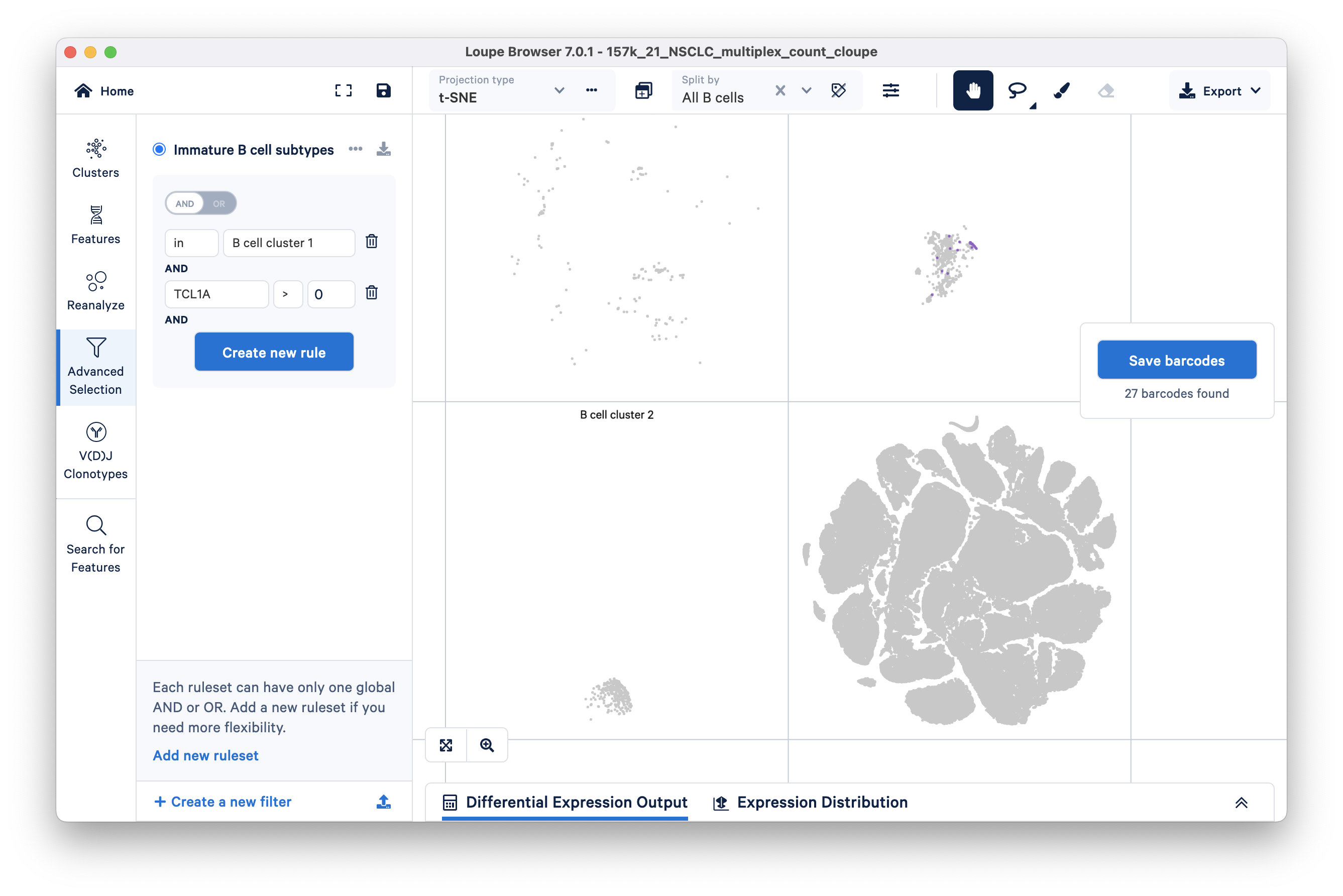
TCL1A gene is expressed in 27 barcodes within B cell cluster 1.
To find immature B cells in cluster 2, click the 'Add new ruleset' option at the bottom of the panel. Add a cluster rule and a feature rule similar to the previous step. For this ruleset, the cluster name should be 'B cell cluster 2'.
Finally, switch the ruleset toggle (top toggle) to OR. You will see that 120 barcodes have passed your filtration criteria.
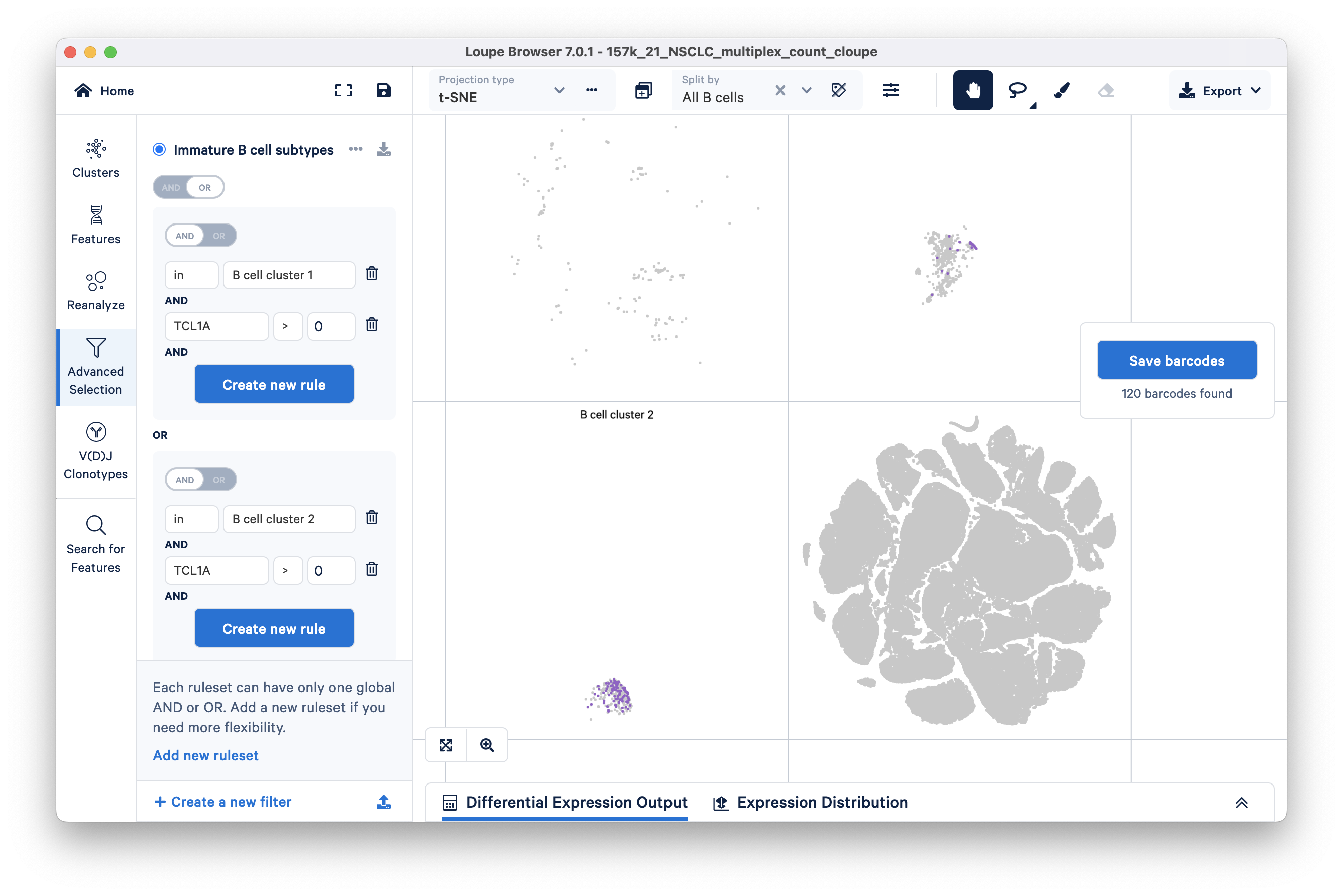
Click the blue 'Save barcodes' button on the right. Name the selected cluster 'immature' and add it to a new group called 'B cell subtypes'. Clicking the Finish button should take you to Clusters mode where you will see a new custom group called B cell subtypes.
For this tutorial, we will classify all barcodes that do not express the TCL1A gene as mature B cells. To filter for mature B cells, modify the existing TCL1A rule so that the rule reads "TCL1A = 0" instead of "TCL1A > 0". You can do so by clicking the greater than symbol (>) directly and choosing equals (=) instead.
Alternatively, you can create a new filter for mature B cells by clicking the 'Add new filter' option at the bottom of the Advanced Selection mode panel. The primary advantage of creating a new filter (over modifying an existing one) is that you can save your filtration workflow for future reference.
Your filter should look like this:
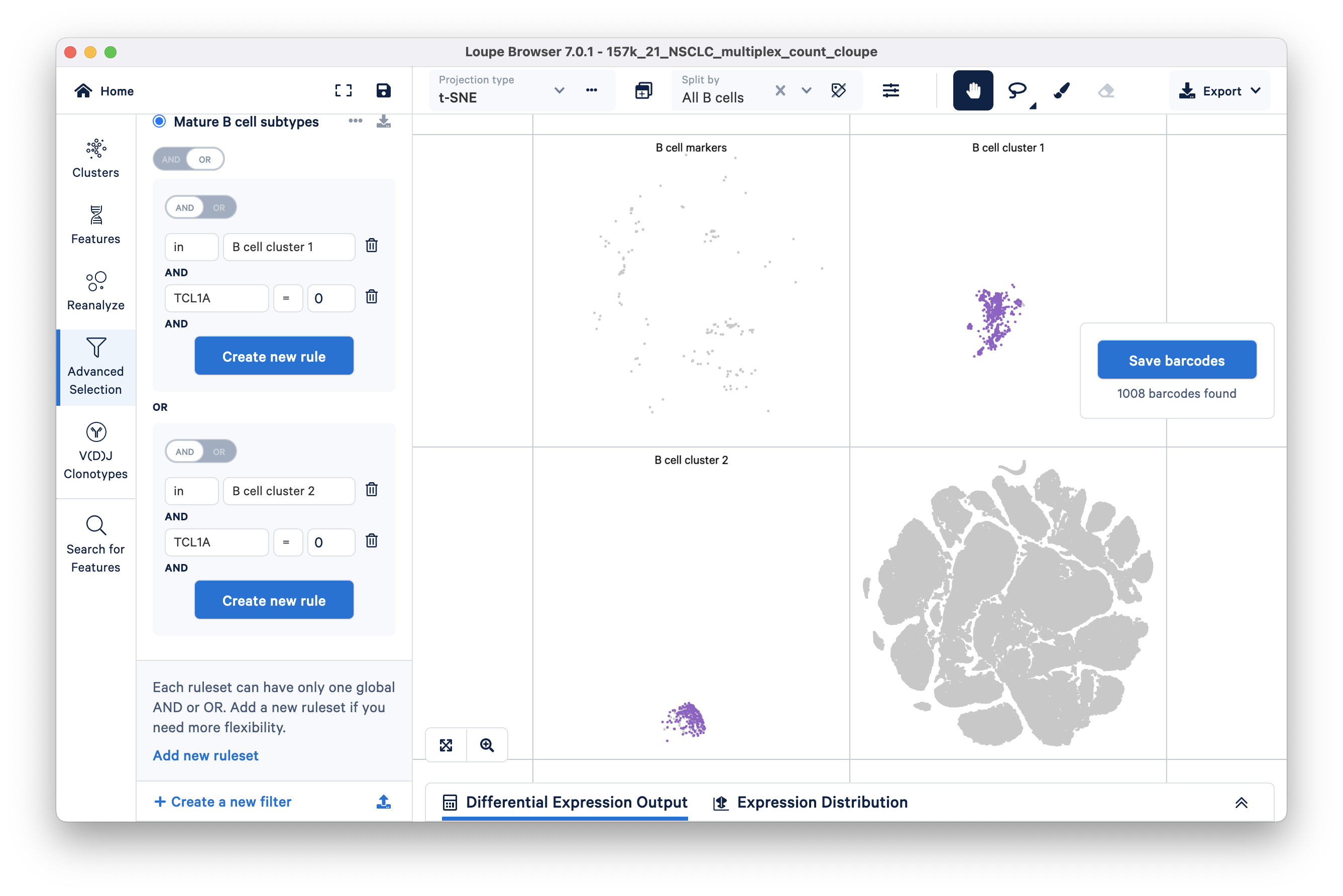
A different subset of B cells should now be highlighted in purple. Assign these 1008 barcodes to the same category "B cell subtypes" and name the cluster "mature".
Now that the B cells in this dataset are segmented into mature and immature, you can choose to run differential expression between these subtypes (discussed in the Navigation and Differential Gene Expression Analysis tutorial). It is interesting to note that both clusters are primarily composed of mature B cells. However, cluster 2 has more immature B cells than cluster 1.