Note: 10x Genomics does not provide support for community-developed tools and makes no guarantees regarding their function or performance. Please contact tool developers with any questions. If you have feedback about Analysis Guides, please email analysis-guides@10xgenomics.com.
The 10x Genomics Chromium Single Cell platform facilitates the generation of single cell CRISPR screen data, where CRISPR perturbations of genomic elements are delivered to cells and measured alongside whole transcriptome gene expression or other molecular phenotypes. Such data have immense potential for functional genomics discovery but present several statistical and computational challenges. Our Cell Ranger pipeline enables convenient analysis of CRISPR screen data. However, if you seek greater analysis flexibility or visualization capabilities, and better scalability to large CRISPR screen data, community-developed tools, such as sceptre, could be helpful for you.
sceptre is an R package for single cell CRISPR screen analysis that emphasizes statistical rigor, computational efficiency, and ease of use. This post is intended to serve as an introduction to sceptre. For comprehensive guidance on using the software, please consult the sceptre website and manual.
The sceptre workflow consists of several distinct steps (illustrated below). To use sceptre on 10x data, you will need to first run Cell Ranger to generate count matrices. Guided tutorials and relevant resources are available on the Cell Ranger support website. One key output file from Cell Ranger is the feature-barcode matrix, which is used as the input to sceptre.
After importing your data into sceptre, the tool guides you through a series of steps culminating in a set of element-gene pairs. These pairs represent instances for which perturbation of the genomic element is confidently linked to a change in the expression of the gene. Along the way, sceptre assigns CRISPR gRNAs to cells, runs quality control, and validates the accuracy of the statistical analysis using your designated positive and negative control perturbations. At each stage, sceptre provides status updates and informative plots to monitor the progress of your analysis.
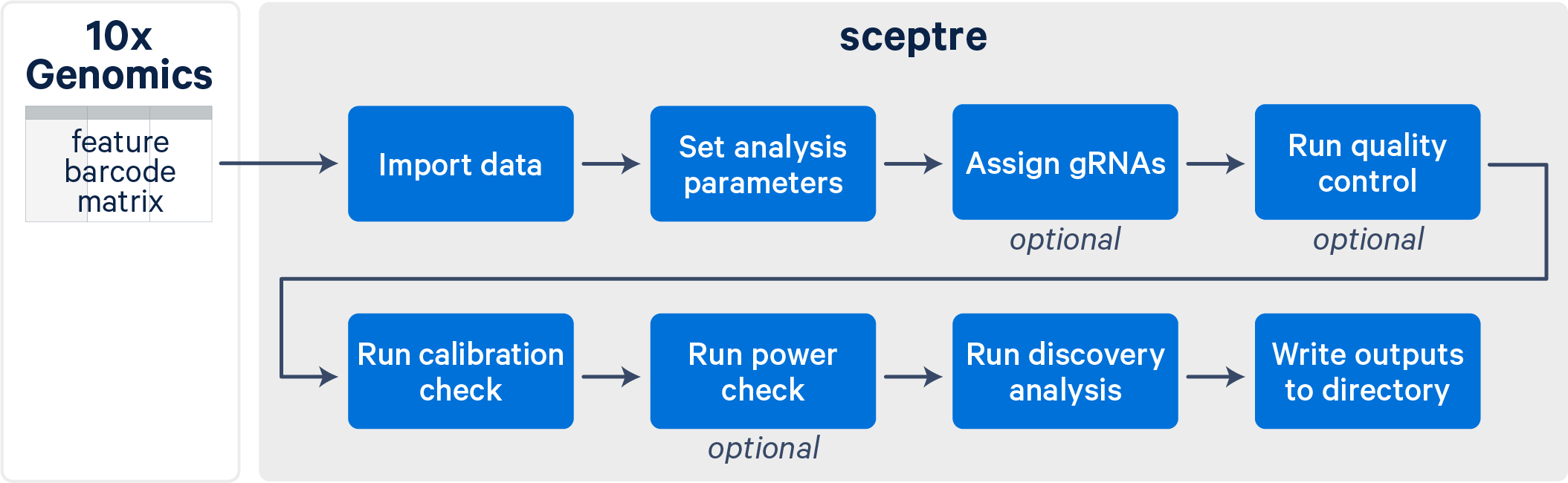
Users of sceptre enjoy access to a range of features and options, a large and reliable set of biological discoveries, low memory usage and compute time, and a smooth user experience.
A range of features and options
Single cell CRISPR screening involves several key experimental design choices, such as the number of CRISPR perturbations introduced per cell (single or multiple), target region (coding or noncoding elements), and the CRISPR modality used to affect the perturbation (CRISPRko or CRISPRi). sceptre supports these diverse experimental combinations with a comprehensive analysis toolkit. Beyond its core element-gene analysis functionality, sceptre offers functions to construct element-gene pairs to test for association. For example, construct_cis_pairs() and construct_trans_pairs() are useful in the context of screens perturbing noncoding regulatory elements and genes, respectively.
Many reliable biological discoveries
At the core of the sceptre software is the SCEPTRE method [1, 2, 3], which has been demonstrated to provide state-of-the-art sensitivity and specificity in testing element-gene pairs for association. This method combines a negative binomial model with a resampling framework for statistical inference, offering a significant improvement over traditional negative binomial models. In practice this enables the identification of a greater number of more biologically meaningful element-gene links. Importantly, sceptre allows users to verify the quality of the analysis on their data via its calibration check and power check functionalities. If there is any evidence of miscalibration, Chapters 5.3-5.4 of the manual provide guidance on diagnosing and correcting these issues.
Computational efficiency
Despite the high computational demands of resampling-based methodologies, the sceptre software is fast due to extensive optimization. In a recent benchmarking analysis on a dataset with 29,000 cells, sceptre demonstrated remarkable efficiency, analyzing over 200 element-gene pairs per second. During the calibration check, which lends itself to even further acceleration, its speed approached 10,000 element-gene pairs per second. Besides computational speed, sceptre is also designed to be memory-efficient. The memory required to analyze a dataset is typically less than twice the size of the dataset itself (when stored in standard sparse R matrix format).
User experience
sceptre simplifies the analysis of single cell CRISPR screen data with an intuitive object-oriented interface. The data are stored in an R object called a sceptre_object. At each stage of the pipeline, plot(sceptre_object) produces relevant visuals, while print(sceptre_object) produces helpful textual summaries and progress updates. The software conducts extensive input validation and prints clear error messages if an input is incorrectly specified. Comprehensive documentation offers detailed guidance on the analysis choices available at each step of the pipeline, a glossary of key terms, and a chapter on FAQs makes the entire process straightforward and user-friendly.
Visit the official sceptre website and online user manual for download instructions and examples of analyzing single cell CRISPR screen data. To engage with the sceptre developers and community, open an issue in the sceptre Github repository to report a bug, start a discussion to request a feature, or ask a question about the software. If you do not wish to post your question publicly, email the principal investigator or lead developer of the software.
References
[1] T Barry et al., SCEPTRE improves calibration and sensitivity in single-cell CRISPR screen analysis. Genome Biology (2021). doi: https://doi.org/10.1186/s13059-021-02545-2
[2] T Barry et al., Exponential family measurement error models for single-cell CRISPR screens. Biostatistics (2024). doi: https://doi.org/10.1093/biostatistics/kxae010
[3] T Barry et al., Robust differential expression testing for single-cell CRISPR screens. Genome Biology (2024). doi: https://doi.org/10.1186/s13059-024-03254-2