Exploring cancer progression and therapeutic outcomes with multiomics: Discussions from the 2021 Global Virtual Cancer Symposium
This year’s Global Virtual Cancer Symposium let us learn about some of the latest discoveries in cancer biology, with leading cancer researchers sharing their work to better understand cancer progression and therapeutic response and resistance. Take a look at their exciting research, and get started on your own with our new sample prep guide for single cell applications in oncology.
We are continually impressed by the research being done in oncology, as scientists work tirelessly to reveal the underlying mechanisms in cancer progression. Five leading researchers have put cutting-edge multiomic technologies to the test, improving our understanding of cancer biology and pointing us in the direction of new therapeutic targets. We were thrilled to hear them share about the advances they’ve made and the methods that made them possible at our recent Global Virtual Cancer Symposium.
We hope you were able to join us, to hear about the exciting work being done and to participate in the Q&A sessions with the researchers themselves, as they fielded questions about the considerations involved in their current approaches and how they see their research moving forward. If you were unable to attend, don’t worry! Each talk is available in its entirety—and that includes the Q&A discussions, too. Read on for highlights from the presentations and access to the on-demand video series.
The roles of tissue of origin, proliferation, and differentiation in CD8+ T cell responses to lung cancer
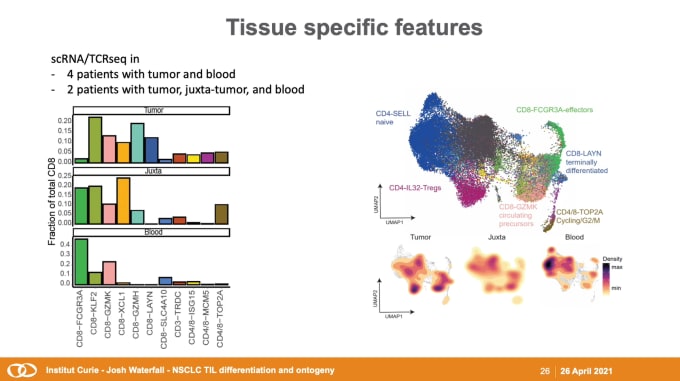
Josh Waterfall,PhD, Team Leader for the Integrative Functional Genomics of Cancer at Institut Curie, got the week started, diving into the dynamics between tumors and the immune system. His team of computational genomicists collaborates closely with colleagues in chromatin biology and immunology to investigate the interactions between tumors and the TME. He discussed their recent studies on non-small cell lung cancer (NSCLC), the leading cause of cancer-associated death worldwide. As an immunogenic tumor type, tumor-infiltrating lymphocyte (TIL) invasion is a positive marker for prognosis, and Dr. Waterfall’s team analyzed single cell RNA-sequencing (scRNA-seq) and T-cell receptor (scTCR) repertoire profiling to identify variation in TIL subpopulations across NSCLC patients and tissue types. This data feeds into the development of a comprehensive model for T-cell differentiation and dysfunction, with the goal of improving clinical care for cancer patients.
Spatial and single-cell transcriptomics analysis of neoadjuvant anti-PD-1 therapy in hepatocellular carcinoma
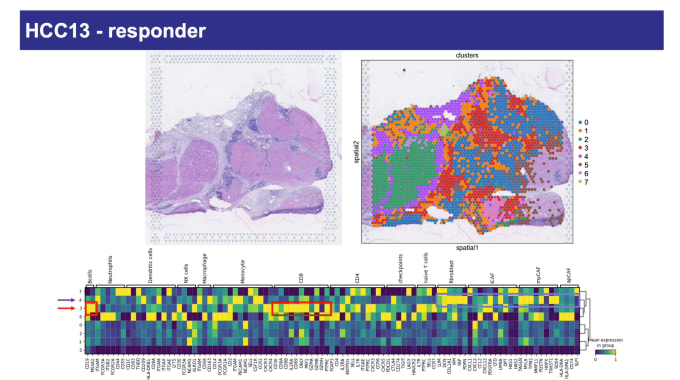
Luciane Kagohara, PhD, Assistant Professor of Oncology at Johns Hopkins Medicine, talked to us about her work in improving treatment outcomes for patients with hepatocellular carcinoma (HCC), the most frequent type of liver cancer. At the time of diagnosis, surgery is not a viable option for the large majority of patients, and even those who have surgery usually see the cancer recur. Dr. Kagohara’s team is investigating the effectiveness of neoadjuvant anti-PD-1 therapy as a treatment option. They performed single cell immune profiling and spatial gene expression analysis to identify the transcriptional signatures of response and resistance to this immunotherapy in a phase 1 clinical trial. Spatial analysis proved to be a particularly powerful tool for visualizing features associated with anti-PD-1 treatment. Those responding positively to treatment showed clusters of cytotoxic T cells and other immune populations in close proximity to tumor regions. For non-responders, while immune cells did infiltrate tumors, they did not form aggregates. Altogether, these data indicate probable patterns connected to clinical response.
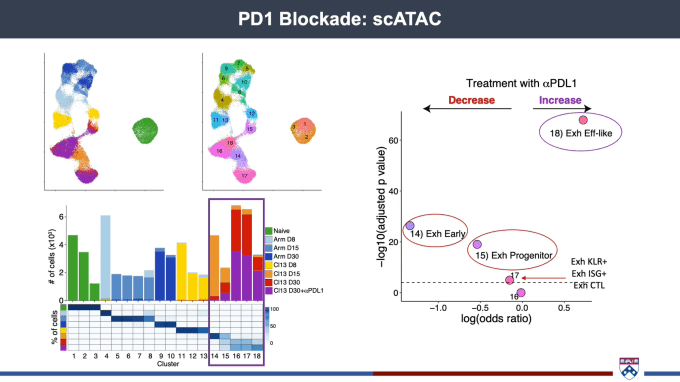
T cell differentiation at the single cell level: Implications for T cell exhaustion and immunotherapy
Josephine Giles, PhD, Mark Foundation Cancer Research Institute Postdoctoral Fellow at the University of Pennsylvania, spoke about the importance of getting as granular and detailed a picture of the spectrum of T-cell fates as possible. Most cancer patients do not achieve a durable clinical response after immunotherapy, suggesting that treatment is not producing a robust pool of healthy effector cells. With this in mind, her team has been working to gain a deeper understanding of T-cell differentiation and function. scRNA-seq allowed them to capture the complexity inherent in the transition from effector to memory T cells, including transcriptional diversity within exhausted T-cell populations. Turning to chromatin accessibility to better identify cell type, Dr. Giles’ team performed scATAC-seq on the same system and found similar cellular subsets as had been defined by scRNA-seq. With epigenetically distinct subsets of exhausted T cells, part of the struggle in turning them into healthy effectors may lie in the need for reprogramming to take place at the epigenetic level to allow for differentiation.
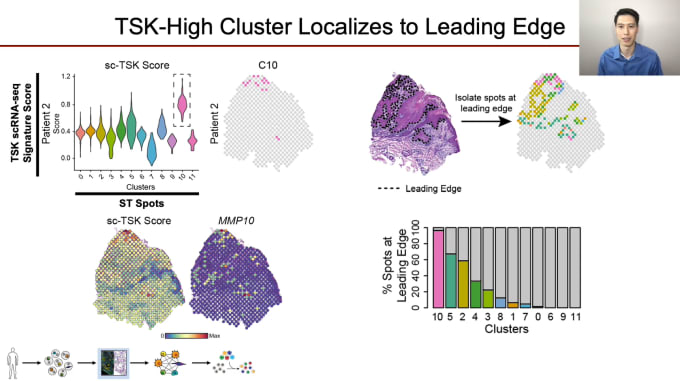
Single-cell and spatial dissection of cellular communication in squamous cell carcinoma
Andrew Ji, PhD, Clinical Instructor and Postdoctoral Fellow of Dermatology at Stanford University, is proving just how critical a role cellular communication plays in cancer progression and treatment. His team is looking closely at how crosstalk between various cell types with tumors leads to intratumoral heterogeneity (ITH). Development of ITH is a broad issue across all cancers, leading to treatment resistance. Dr. Ji’s work focuses on cutaneous squamous cell carcinoma (cSCC), the second most common cancer overall in the US. While most cases are cured with excision, about 4% develop lymph node metastasis, at which point treatment is quite difficult. In studying cellular communication in cSCC, Dr. Ji and his colleagues integrated scRNA-seq data with high-resolution spatial profiling to fully characterize the organization of the tumor and TME, and identify a novel subpopulation of cells with the potential to act as a source for crosstalk at the leading edge. This new subtype may well serve as a future therapeutic target, and as cSCC shares cardinal features with other epithelial cancers, knowledge gained here could possibly be applied more broadly.
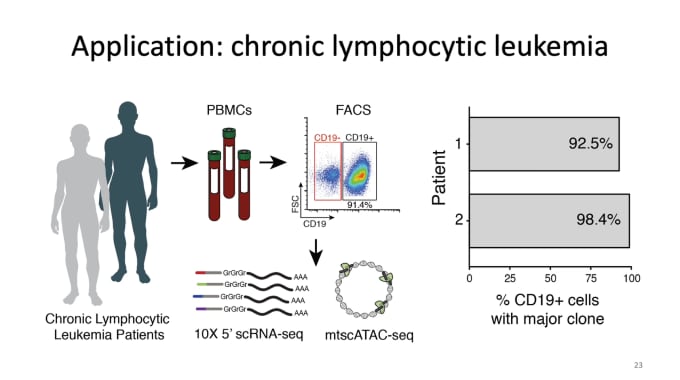
Clonal lineage tracing in healthy and malignant human tissue via single-cell genomics
Caleb Lareau, PhD, a Postdoctoral Research Fellow in Pathology at Stanford University, closed out the week by sharing his work in lineage tracing in humans. Understanding clonal evolution goes hand in hand with understanding tumor evolution, with clonal heterogeneity in tumors likely playing essential roles in cancer progression, variability in treatment response, as well as relapse. With this motivation, Dr. Lareau’s team focuses on deconstructing these polyclonal populations via identification of somatic mutations in mitochondrial DNA. In a truly innovative move, they made a few key modifications to our scATAC-seq protocol that would allow them to retain mitochondria during sample prep and perform mtDNA sequencing at greatly increased throughput levels, and then developed a new computational framework for analysis. In applying this new protocol to chronic lymphocytic leukemia and colorectal cancer, they could identify hundreds of somatic mutations in peripheral blood and dozens of mutations to determine potentially functional subclonal features of human tumors. Success with this method led them to go even further, layering on additional modalities, including protein and RNA, for more comprehensive studies.
With so much inspiring work being done with single cell multiomics, scientists are paving the way for future research and potential breakthroughs. We can’t wait to see what the coming years bring. And that includes you—how are you planning to move forward with your own cancer research? If you’re looking to incorporate single cell applications into your studies, we’re here to help you get started. Even if you’re new to single cell technology, the first step is one familiar to biologists the world over: sample preparation. With that in mind, we’ve created a new resource to help you get the best possible start.
In this sample preparation guide, you can explore technical notes, protocols, and more for general guidance with single cell applications for oncology as well as concerns specific to your tissue type. A curated list of publications categorized by cancer type makes it easy to learn from researchers who have traveled similar paths before.