From one gene to thousands: Scaling spatial genomics efforts with high-throughput single cell spatial imaging
Impact at a glance: Dr. Kenny Roberts, Senior Staff Scientist at the Wellcome Sanger Institute, was doing 150-plex manual in situ sequencing experiments. Now he’s taking advantage of the Xenium In Situ platform to study hundreds (soon to be thousands) more transcripts, accelerate experiments from two weeks to two days, see data faster with onboard analysis, and get reliable results across sample types with reproducible standardized protocols.
“If I'd known then that we'd be doing 400-plex in situ in an automated fashion and that on the horizon we’d potentially have 5,000-plex, I almost wouldn’t have believed it.”
The mission of Wellcome Sanger Institute’s High Throughput Spatial Genomics (HTSG) Initiative is monumental. A collaborative effort of both faculty teams and centralized scientific pipelines, they hope to realize the promise of spatial transcriptomics technologies to build 3-dimensional cellular atlases of entire tissues/organisms and analyze hundreds of patient samples. We recently spoke with Kenny Roberts, PhD, a Senior Staff Scientist in the HTSG group about how single cell spatial imaging with Xenium will help them towards their mission of generating spatial transcriptomic data at scale for the projects that they serve.
Could you please describe your current research projects?
I work within a team that is all about spatial genomics. Our remit really is to provide cutting edge spatial data to other people's biological projects. Obviously, here at Sanger, we have faculty teams working on all manner of different things. Locally, our program (Cellular Genetics) is very heavily involved in the Human Cell Atlas and cell atlasing of lots of different human tissues throughout all different life stages. We provide spatial data across both our program and, more recently, other teams across the institute.
For example, at the moment, we're finishing a project on Hox gene mapping across the developing human spinal cord led by Dr. Sid Lawrence and Dr. Sam Behjati. We've also been doing some mapping of autism gene expression across the developing human brain in the second trimester with Dr. Omer Bayraktar, who's my team leader. And with one of the clinical fellows in our Cancer, Ageing and Somatic Mutation program, Dr. Lucy Yates, we're looking at cell atlassing across different types of breast cancer ranging from early-stage ductal carcinoma in situ through to the more invasive infiltrating ductal carcinoma.
How did you become interested in studying spatial biology? Did it happen serendipitously, or did you know going into research that you really wanted to focus on this?
I think spatial biology sits at a confluence of different interests that I have, but there was also a lot of serendipity. I studied biochemistry as an undergrad, then my PhD was very molecular biology heavy and I really grew to enjoy imaging at that time. Actually, I remember one of my supervisors, during a hard time saying, “Plan some imaging because it always cheers you up.” Even now after all the thousands of images I've generated, I love just seeing images as they come off the microscope.
So, in my PhD, I worked for four years on one gene, which felt like I was in a very small niche. Then, when I saw this job advertised at Sanger in spatial transcriptomics, it just seemed like a great opportunity to take the imaging and molecular biology skills to a bigger scope. Last year I gave a career development talk, and I had a slide showing how I went from looking at one gene in one tissue to now looking at 100 to 20,000 genes in more than 25 different tissues.
What led you to consider adopting newer single cell spatial imaging technology for the projects you are working on?
Since I've been at Sanger, we've gradually been increasing the complexity of the spatial technology we've been doing. When I first started, we did a lot of four-plex single-molecule fluorescence in situ hybridization (smFISH), lots of antibody staining, and it was enough to answer some of the basic questions to validate some of the main findings in our single cell data, but we weren't able to do this sort of full-scale atlassing that I've just mentioned.
As we've moved into (pre-Xenium) in situ sequencing (ISS) we've finally been able to really start mapping the cell types of different tissues. The downside is the sheer amount of time and effort that it requires to undertake those experiments. We're looking at the better part of two weeks for a slide of manual in situ sequencing. The data's been fantastic, but the amount of time means we can't really hit that high-throughput level that we would like to. We can't equitably provide that to many teams across the institute all at once. We need a more refined way of generating that data and going beyond the 150-plex we were doing manually. Xenium in particular has been a real advantage for us with that.
It sounds like there were a lot of technical limitations with manual in situ sequencing and some of the workflow benefits of Xenium are what led you to choose it. What other elements of Xenium have been beneficial to your high-throughput spatial genomics initiative?
Having reproducible standardized protocols that will work for everything is super advantageous. We work with all of these different types of samples. We have frozen tissue and FFPE and other preparations that we'd like to be able to work with as well—all sorts of different tissues from different life stages in humans and, potentially, other species—so the applicability to different samples is incredibly useful.
The streamlined workflow is automated so that we can now go from this protocol that was taking two weeks of manual work to two days of work. Now, the most tedious, repetitive [part]—and, actually, the part of the manual protocol where I found things used to go wrong—is replaced by a standardized protocol and being run on an instrument in an unsupervised way.
Also, as the name of our team suggests, we want something that's high throughput. From the systems that we evaluated, Xenium certainly seems to be the fastest at providing this data, and that was an important consideration for us.
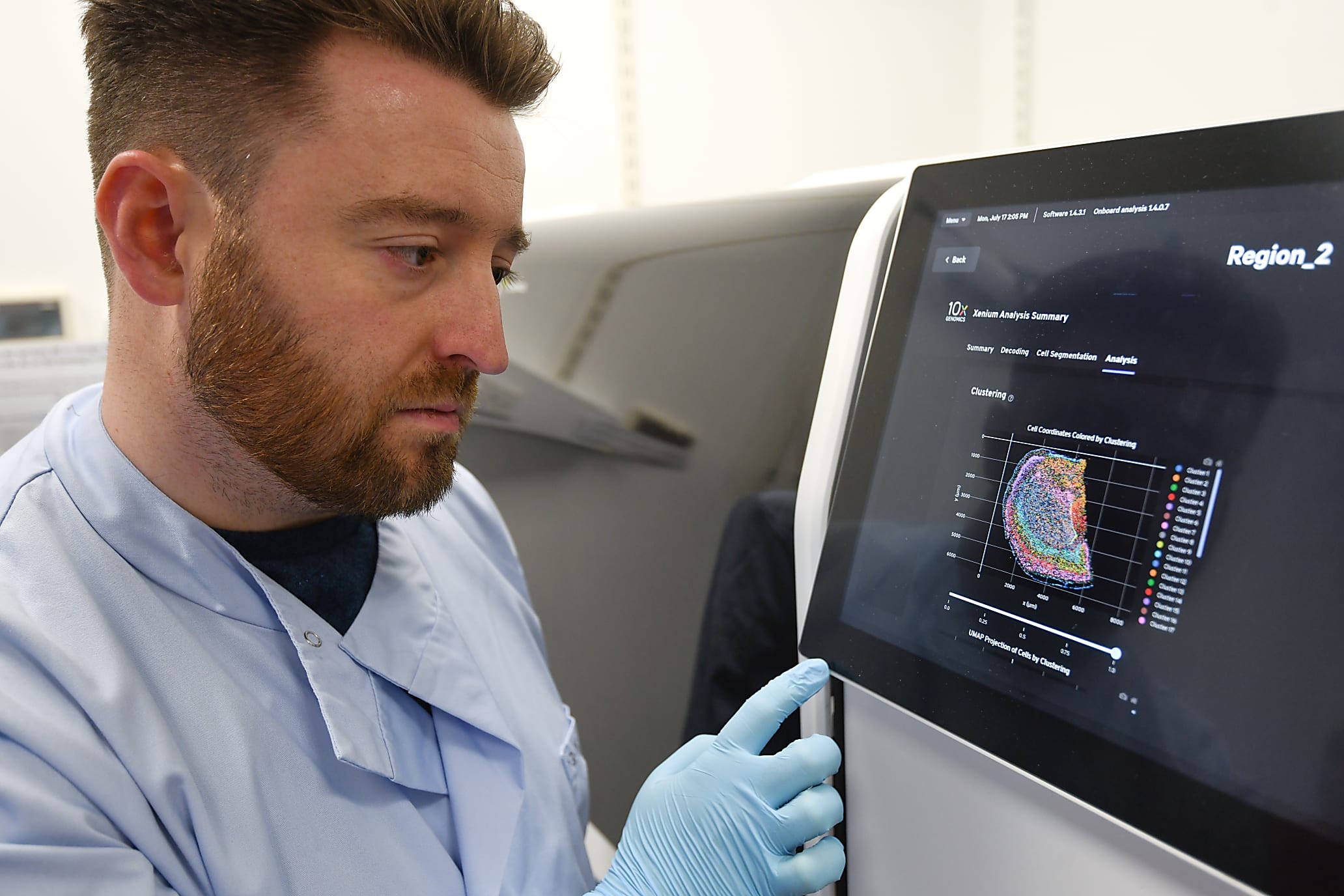
Finally, for me, as a wet lab biologist that doesn’t really do any computational analysis, the idea that the instrument does a huge amount of the analysis on board and gives me immediately processed data when the run is finished is a huge advantage. I've been very lucky to work with some great computational people here, particularly our senior software developer, Dr. Tong Li, who has led all of our image wrangling and ISS decoding for our other projects. But to be able to go to the instrument and immediately see the data, be able to download it, and be looking at it within minutes rather than a process that used to take weeks with a huge amount of computational resources and time is just fantastic. It is so exciting to me that I can see the data myself that I've prepared.
How will having access to Xenium help shape your group moving forward?
So I think certainly we should be able to provide this level of data to more collaborations. A lot of the projects that we work with would benefit from this kind of data, but the pragmatic balance of what we could feasibly achieve with previous iterations of ISS meant that there were a limited number of projects we could actually apply it to. Now, we really are looking at being able to pipeline in a much higher-throughput way, so that people can bring us projects and we can sort of turn them around with much more agility than we could before.
I think, also, it's drawing attention from people that previously weren't that interested in spatial or didn't necessarily see the value of it. The data we've already generated is having greater visibility to other teams that are seeing it and going, “Oh, this looks really cool. We'd actually really benefit from this. We'd really love to have this for this project that at the moment we don't have spatial data for.”
Speaking of exciting data, can you talk about the experience of seeing your first dataset?
I can definitely say that I've been super excited every single time I've seen the instrument finish a run, and that's also true of all of the collaborators we provide data for. I think people are just completely blown away by it, particularly those that have not seen spatial data before. In some ways, I think when you understand what a leap Xenium has made, you almost appreciate it more, especially the people that we were previously working with, doing manual ISS, who were waiting months every time for us to just do a little bit of data. For them, now, to receive it in a format that they can view themselves in the Explorer software, they can really interrogate it almost immediately after we've discussed the experiment—people are just absolutely mind blown.
Any closing thoughts?
I read something interesting the other day that makes for a good comparison when I think about how this technology will shape the field going forward. In this article, our Director of Scientific Operations, Dr. Cordelia Langford, did an interview with the Genetic Society Podcast. She was talking about the history of Sanger and DNA sequencing, and her memories of joining Sanger nearly thirty years ago. They were making the transition from doing these autoradiographs of gels for DNA sequencing where you would read off one base at a time on the gel to the precursors of what are now our sequencing platforms. She was saying that it was mind-blowing—the same word I keep using about in situ sequencing and Xenium—to sequence in this automated way using these capillary sequencers. Looking back now, they were only doing a few hundred bases at a time, which now seems like nothing, but at the time, it was amazing.
If they'd known then what we can do now, it would have just been unbelievable. That really is how I feel about my experience with spatial over the last five years. When I was doing my PhD, I did one-plex chromogenic in situ hybridization experiments with DIG-labeled probes and I even thought that was amazing to be able to see the transcripts showing up on the tissue. Then, coming to Sanger and doing three- or four-plex FISH, I thought that was the best thing ever. But, if I'd known then that we'd be doing 400-plex in situ in an automated fashion and that on the horizon we’d potentially have 5,000-plex, I almost wouldn’t have believed it. I think it's just a really interesting parallel—separated by three decades, but it's almost the same process of scientific development.
This interview has been edited for length and clarity.
To learn more about Dr. Roberts’ work, visit the Wellcome Sanger Institute’s High Throughput Spatial Genomics website. Read more about the Xenium platform, here.
