Spatial transcriptomics publications you’ll want to read
Have you heard the news? Back in January, Nature Methods announced spatially resolved transcriptomics was its 2020 Method of the Year, and we were absolutely thrilled. Judging by the responses to their Twitter announcement, we weren’t the only ones.
The increasing number of preprints and publications featuring spatial transcriptomics highlights the growing popularity and utility of spatial profiling methodologies. In fact, at the time of writing this blog, a search of the bioRxiv preprint archive for the term “spatial transcriptomics” yielded 480 results, 70 of which were submitted in 2021.
With so many papers available to read and not enough time to read them all, where is a researcher to start? We’ve got you covered with a list of some of our favorite recent publications featuring RNA seq–based spatially resolved transcriptomics.
Building a human intestine
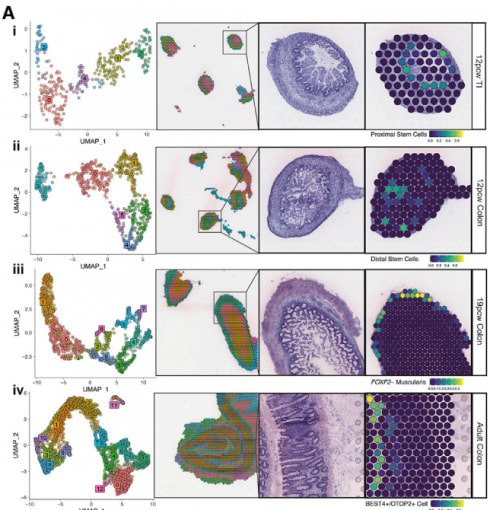
Species-specific developmental differences often make it difficult to interpret how findings from animal models correlate to human development. To get a better understanding of human intestinal development, Fawkner-Corbett et al. integrated spatial transcriptomics and single cell RNA-seq (scRNA-seq) data, generated using Visium Spatial Gene Expression and Chromium Single Cell Gene Expression, to map morphogenesis in embryonic samples over time and across location, even down to the cellular compartment level (1). Promisingly, the spatial gene expression data showed a layered, ring-like distribution that suggests tissue depth plays an important role in determining transcriptional/cellular spatial variability. Using this two-pronged approach, the researchers observed the coordinated emergence of different cell populations, defined the regulating transcription factor networks, and charted intercellular communication.
Generating a timeline of sepsis pathogenesis
The Centers for Disease Control and Prevention estimates that roughly 1.7 million Americans develop sepsis each year, with as many as 270,000 cases becoming fatal (2). Given that acute kidney injury is a common and often fatal complication of sepsis, Janosevic et al. sought to understand the pathophysiology of kidney injury using a murine endotoxemia model of sepsis (3). Spatial transcriptomics analysis of endotoxemic mouse kidneys, generated with Visium Spatial Gene Expression, helped pinpoint the location of a unique cluster of proximal tubular cells the study identified via scRNA-seq. The authors suspected this cluster represented proximal tubular S3-Type 2 cells—identified by other studies—localized to the outer stripe of the outer medulla, whose microenvironment may explain its unique transcriptional signature compared to other proximal tubular cells. The authors note that,
“Our work points to the urgent need for defining a more accurate and precise human sepsis timeline. Such definition will guide the development of biomarkers and therapies that are cell and time specific.”
Understanding schizophrenia at the cellular and tissue levels
Schizophrenia is an extremely complex neurological disorder that impacts a wide range of cognitive functions, from memory to language to emotions, yet its pathogenesis is poorly understood (4). To understand which neuronal subtypes are most affected in patients with schizophrenia, Batiuk et al. (in preprint) used a combination of single nucleus RNA-sequencing (snRNA-seq) and spatial transcriptomics to profile transcriptional states of neurons within a region of the dorsolateral prefrontal cortex (DLPFC) that has been linked to functional, genomic, and morphological abnormalities. Visium Spatial Gene Expression helped the researchers visualize the upper, or supragranular, layers of the cortex as a hotspot for transcriptional and compositional changes associated with schizophrenia. Altogether, this preliminary data points to a widespread impairment of multiple neuronal subtypes within the supragranular layers of the cortex.
Mapping tumor interactions with new microenvironments during invasion
To successfully spread, tumor cells must interact with cells in the new environments they seek to invade. While single cell sequencing has greatly increased our understanding of tumor heterogeneity, there is a loss of spatial information that limits a detailed understanding of the tumor interactions with its microenvironment (TME). Taking advantage of a technique that integrates spatial transcriptomics and scRNA-seq data, Hunter et al. set out to characterize the intercellular interactions occurring at tumor/TME borders in a zebrafish melanoma model at cellular resolution. Uniquely, because adult zebrafish measure roughly 5 mm in diameter, an entire transverse section fits within a single Capture Area on a Visium Spatial Gene Expression slide. This allowed the researchers to study the tumor and all surrounding tissues intact. The researchers identified a transcriptionally unique “interface” cluster at the border between tumors and TME, despite no visible morphological differences. Further analysis revealed that this interface is composed of specialized tumor and muscle cells, with a unique “interface” transcriptional state, that coordinate to promote gene expression programs that mediate tumor invasion.
The exciting future for spatial transcriptomics
These are just a sampling of the many exciting and groundbreaking applications of spatially resolved transcriptomics. We are eager to see the additional breakthroughs that spatial transcriptomics will lead to as methodology improvements open up its use to FFPE tissues and allow single cell resolution.
If you want to learn more about Visium Spatial Gene Expression for whole transcriptome mapping in morphological context, contact us or visit: https://pages.10xgenomics.com/visium-breaking-spatial-barriers.html.
References
- Fawkner-Corbett D, et al. Spatiotemporal analysis of human intestinal development at single-cell resolution. Cell 184(3):810-826.e23, 2021. doi: 10.1016/j.cell.2020.12.016
- Clinical information. (2020, December 07). Retrieved March 12, 2021, from https://www.cdc.gov/sepsis/clinicaltools
- Janosevic D, et al. The orchestrated cellular and molecular responses of the kidney to endotoxin define a precise sepsis timeline. eLife 10:e62270, 2021. doi: 10.7554/eLife.62270
- Schizophrenia. (2019, October 4). Retrieved March 12, 2021, from https://www.who.int/news-room/fact-sheets/detail/schizophrenia
- Batiuk MY, et al. Selective vulnerability of supragranular layer neurons in schizophrenia. bioRxiv, 2021. doi: 10.1101/2020.11.17.386458
- Hunter MV, Moncada R, Weiss JM, Yanai I, and White RM. Spatial transcriptomics reveals the architecture of the tumor/microenvironment interface. bioRvix, 2021. doi: doi.org/10.1101/2020.11.05.368753
