Untangling the cellular and molecular drivers of neurodegeneration: A look back at the Global Neurodegenerative Disease Summit
An “invisible pandemic”—that’s how Dr. Nancy Ip, Vice President for Research and Development and Morningside Professor of Life Science at The Hong Kong University of Science and Technology, described Alzheimer’s disease, one of the most common forms of neurodegenerative disease, at our Global Neurodegenerative Disease Summit. And it makes a lot of sense, especially when you look at the numbers.
During the second half of the 20th century, we saw a massive increase in global life expectancies, but this rise in longevity has turned out to be a double-edged sword. As we age, the likelihood of developing neurodegenerative disease increases significantly. According to the Society for Neuroscience, there are an estimated 50 million people worldwide suffering from some form of dementia. By 2030, this number will likely reach tens to hundreds of millions.
That’s why we chose to highlight some of the brightest minds in neurodegenerative disease research during our recent summit, inviting leading neuroscientists to present their latest findings. We got an inside look at some of their fascinating work and saw first-hand how they’re using single cell and spatial technologies to untangle the cellular and molecular drivers of neurodegenerative disease.
Keep reading for a quick rundown of our Global Neurodegenerative Disease Summit, then get into the details by watching the presentations—now available on-demand—for yourself.
And the Early Career Investigator Award goes to…

Every single neuroscientist is looking toward a future without neurodegenerative disease. No one can predict how far off that may be, but with new discoveries and new researchers entering the field all the time, the promise of that future grows. We started off the Global Neurodegenerative Disease Summit with this hope in mind, announcing the winner of our Early Career Investigator Award, Dr. David Gate, Assistant Professor at Northwestern University. Focusing on the role of the adaptive immune system in neurodegenerative disease, Dr. Gate’s research has explored the T-cell contribution to Alzheimer’s disease (AD), Parkinson’s disease (PD), and Lewy body dementia (LBD) pathology. With his awarded $35,000 in 10x Genomics products, Dr. Gate plans to extend this research in his lab at Northwestern University. He hopes to further identify T-cell clonotypes—cells that express receptors in both the blood and cerebrospinal fluid—in AD, PD, and ALS utilizing Chromium Single Cell Gene Expression and Immune Profiling. These results will then be studied within the context of disease severity–based patient stratification to investigate whether or not the identified T-cell clonotypes reflect disease pathology.
The good, the bad and the microglia – mechanisms of C9orf72-mediated neurodegeneration
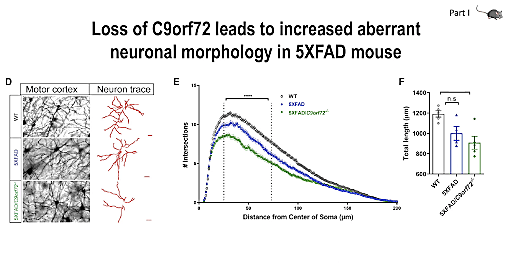
Next, we were joined by our keynote speaker, Dr. Rita Sattler, Professor at the Barrow Neurological Institute and winner of the 10x Genomics 2021 Neuroscience Challenge. With her expertise in preclinical drug development and strong background in translational neuroscience, Dr. Sattler had some exciting insights to share on ALS and frontotemporal dementia (FTD), including the mechanisms through which C9orf72 mutation may contribute to disease development. Dr. Sattler started off with an introduction to ALS and FTD, neurodegenerative diseases representing two opposite ends of a spectrum, exhibiting pathological and genetic overlap. Dr. Sattler’s research focuses on the role of C9orf72 repeat expansion in the context of ALS and FTD, specifically as it pertains to microglial cell function. Though they make up less than 10% of the glial population within the brain, microglia play a critical role in synaptic pruning, which, when dysregulated, can promote damage caused by neurodegenerative insult, leading to loss of synapses—an early symptom of ALS and FTD.
Dr. Sattler and her team are working to better understand the mechanisms driving dysregulation and the resulting synaptic loss and damage. Using single cell RNA-sequencing (scRNA-seq) in C9orf72 KO mouse models, they examined the effects of C9orf72 deficiency in microglial subpopulations. They found that loss of C9orf72 in microglia changed their transcriptional profiles, leading to an increase in synaptic pruning that contributes to neuronal dysfunction. Dr. Sattler later gave us a preview of her as yet unpublished single nuclei RNA-seq (snRNA-seq) data from C9orf72 prefrontal cortex. Ultimately, Dr. Sattler and her team hope that their research will lead to improved patient outcomes, identifying potential targets for early disease treatment, before neurodegeneration has progressed into more damaging later stages.
Spatiotemporal dynamics of molecular pathology in ALS
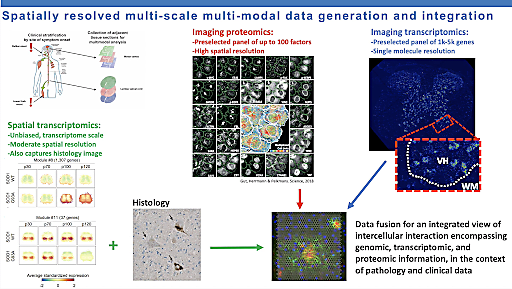
Director of the Center for Genomics of Neurodegenerative Disease at the New York Genome Center, Dr. Hemali Phatnani also presented findings focused on the ALS–FTD clinical spectrum. However, Dr. Phatnani and her team are approaching the problem from a different angle. They aim to understand the spatiotemporal dynamics of ALS and FTD disease progression and how those dynamics are affected by disease-associated mutations. During her presentation, Dr. Phatnani walked us through their process, combining a variety of sample types, including postmortem human brain and mouse models, and techniques, including bulk RNA-seq and spatial transcriptomics, to get an in-depth look at how gene expression changes not only spatiotemporally but also over the course of disease.
Dr. Phatnani explained that spatially resolved transcription was an important part of their research because it let her and her team measure gene expression in multiple cell types and locations simultaneously. ALS and other neurodegenerative diseases involve the pathologies of multiple cell types, and understanding how these cell types interact over the course of disease could yield important insights into disease pathogenesis and progression. Dr. Phatnani and her colleagues hope to identify molecular pathways that change in distinct spatiotemporal ways during disease progression and then use this valuable information as the basis for future therapeutic interventions.
Molecular characterization of selectively vulnerable neurons in Alzheimer's disease
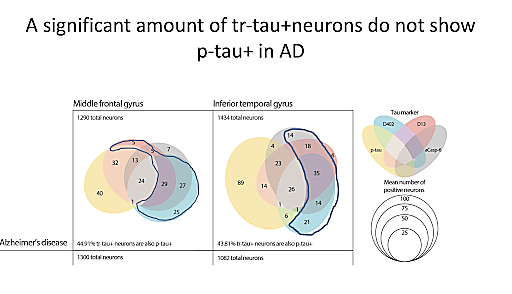
When we think of Alzheimer’s disease, especially from a diagnostic perspective, we generally think of its characteristic accumulation of plaques and tangles, and these aspects are often the focus of research and drug development efforts. However, according to Dr. Lea Grinberg, Professor at UCSF, there is much to be learned from selective neuronal vulnerability. In neurodegenerative diseases, subpopulations of neurons often display significant variability in their levels of susceptibility to abnormal function or death in response to specific types of pathological states or injury, such as amyloid plaques and tau tangles in the context of AD. Dr. Grinberg believes that if we can pinpoint the neurons that are most vulnerable, we can then begin to understand where that vulnerability stems from and develop strategies to protect vulnerable populations from disease.
To this end, Dr. Grinberg and her team are using single cell technologies to study areas of the brain that are particularly affected by AD pathology, specifically the entorhinal cortex and the frontal gyrus, both of which are especially prone to the accumulation of neurofibrillary tangles (tau pathology) during AD. Their aim is to characterize the most vulnerable neurons and the molecular mechanisms underlying this vulnerability. Taking this research to the next step, they have also begun to look at populations of neurons that are vulnerable to tau pathology but do not display tau-related changes. In the future, they plan to further their studies with scRNA-seq, exploring selective vulnerability within the framework of atypical AD.
Single cell epigenomics to reveal causal non-coding variants
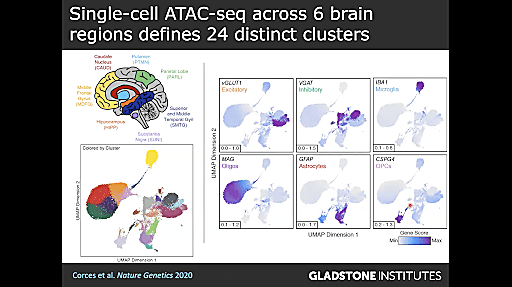
We moved beyond gene expression to epigenetics with Dr. Ryan Corces, Assistant Investigator at the Gladstone Institute of Neurological Disease, whose lab uses a combination of computational biology, large-scale screens, and single cell technologies to explore the contributions of genetic and non-genetic factors to neurodegenerative diseases. Dr. Corces shared his recent studies, discussing his lab’s efforts to understand how the epigenome affects neurodegeneration by annotating the functions of non-coding variants in neurodegenerative disease. They started out with a seemingly straightforward question: why do some people develop neurodegenerative disease, specifically AD and PD, while others do not?
They decided to approach this question from the perspective of inherited genetics of late-onset AD because, though its genetic heritability is 58%, only about 33% of its phenotypic variance is explained by common inherited single nucleotide polymorphisms (SNPs). Based on genome-wide association (GWAS) studies, Dr. Corces and his team knew that much of the remaining phenotypic variance comes from the non-coding genome, but they wanted to understand how these non-coding polymorphisms function and find strategies for identifying them. Using a combination of technologies, including the Chromium Single Cell ATAC (Assay for Transposase Accessible Chromatin) and machine learning, they have been able to leverage chromatin accessibility information to locate epigenomic regions that may affect gene regulatory mechanisms. This multiomic research has the potential to provide new insights into the complicated and multifactorial pathophysiology of neurodegenerative disease as Dr. Corces and his team work to identify these functional noncoding mutations. With this important information, Dr. Corces aims to uncover the unique molecular mechanisms underlying neurodegeneration and reveal potential new genetic targets, improving our ability to predict neurodegenerative disease outcomes.
Single cell transcriptomic analysis reveals the complex cellular mechanisms in Alzheimer’s disease
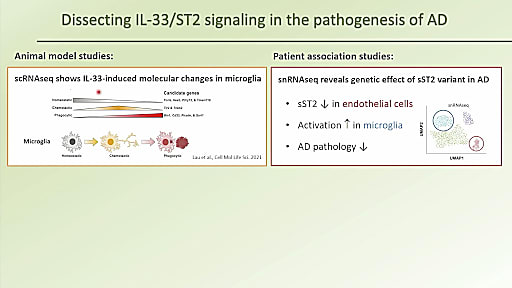
Dr. Nancy Ip concluded our Global Neurodegenerative Disease Summit with her fascinating dive into cell type–specific dysregulation in AD. She began her discussion with a startling truth: there are only six FDA-approved clinical drugs for Alzheimer’s disease and, of those six, five focus on symptomatic relief only. And while the sixth, Aducanumab, targets amyloid, a pathological hallmark of AD, there is a need for disease-modifying therapies that address the molecular causes of AD pathology development. But to develop these new therapeutic interventions, we first need to better understand the mechanisms underlying AD.
With this in mind, Dr. Ip and her team are using single cell and single nuclei RNA-sequencing to take a closer look at two specific cell types implicated in AD—microglia and endothelial cells. In an effort to identify molecules that can regulate microglial activity, they began to study interleukin (IL)-33, an alarmin that is released upon cellular damage in order to maintain homeostasis, conducting a series of experiments with AD mouse models to investigate the effects of IL-33 on AD pathology. With results suggesting that IL-33 has a beneficial effect on AD pathology, specifically in amyloid clearance, Dr. Ip’s team dug even deeper, hoping to understand the pathological factors regulating this process, and identified an IL-33 signaling pathway that is compromised in AD. Using single cell analysis of human samples, they further identified AD-associated genetic variations—driven by an endothelial cell population—that work to modulate AD-related phenotypic traits. Dr. Ip hopes that this research will aid in the development of new potential therapeutic strategies for AD treatment.
Thank you to everyone who joined us at the Global Neurodegenerative Disease Summit and to the amazing researchers who presented their incredible contributions to the field! Watch all the presentations, now available on-demand, here.
