How the VIB Single Cell Core uses Flex Gene Expression to make every experiment feasible
*Impact at a glance: Niels Vandamme, PhD, Head of the VIB Single Cell Core Facility shares how the Flex Gene Expression assay provides operational flexibility and scalability at reasonable costs, making it possible to obtain high-quality data from fragile samples. Plus, explore their unique hack to reduce experimental costs even more.*
“We can, even at the last minute, help them preserve a very, very precious sample and make the study very cost-effective and feasible. We give them feasibility when they don't expect it.” – Niels Vandamme, PhD, Head of the VIB Single Cell Core Facility at Ghent University
This chance to unblock a scientist’s research and make the most of their samples and budget is what excites and rewards the members of the VIB Single Cell Core Facility. Powerful single cell technology, including the Flex Gene Expression assay, and years of hands-on experience are at the heart of this capability.
We sat down with Dr. Niels Vandamme, Head of the VIB Single Cell Core, to speak about how the core onboards researchers and some of the advantages they’ve experienced working with Flex. Keep reading to learn about the operational flexibility they’ve gained through Flex, the robust data quality that keeps them coming back, and the unique way they’re reducing costs for their clients by mixing and matching kitted reagents.
As the head of the VIB Single Cell Core Facility, what factors do you consider when bringing in new technologies?
As a team, we will always respond to user demands, and we will also anticipate user demands. So I think it's very important to have a full future road map of what is going to be accommodated well. We try to identify which will be the long-term technology platforms and technologies that solve current bottlenecks.
When we see technology with the potential to be the next big thing, we act quickly and try to integrate it. And then, once we start, it's about really seeing whether the technology works well.
We have some criteria: it should be robust, reproducible, and scalable. It must be very consistent across tissue types and different labs. We have a multi-site facility, so everybody in the lab should be able to do it in the same way. It should be compatible with our existing infrastructure and our laboratory information management system (LIMS), or we will have to make it compatible with our LIMS and our infrastructure.
I think another factor is whether the new chemistry or assay fits our portfolio of services. Lastly, there has to be a lot of vendor transparency. Good collaboration with the vendor, very strong technical support, and, also, the willingness for core development or beta testing.


What are the most common concerns or considerations of the researchers you work with when it comes to adopting single cell sequencing for their projects?
They usually come with the concern that this is a very complicated logistical workflow. We can easily adjust, adapt, and comfort them because the single cell core is there to work as a kind of one-stop-shop, from the beginning to the end. It comes with a very clear budget overview, with considerations for the full package: for single cell, sorting, sequencing, the data processing, and so on.
Some of the considerations they have at the start of a new project are input requirements and sample integrity. They very often will worry whether they can collect enough cells or if they have enough material or enough nuclei for the project.
A second consideration is usually budget. Can we do the experiment for them within a specific budget constraint or not? It's about balancing your ambitions versus available finances.
Another concern that is quite new in the past years that I've noticed is a lot of awareness around reproducibility issues. People really want to see that the biological relevance is there and that it is also reproducible across multiple experiments.
Additional concerns that we have from users related to reproducibility are: to have no batch effects, to make sure that they have the right experimental design, and that there's no experimental confounders. If they have new assays or chemistries, we, as a core, take responsibility to make sure that for that one big project we have the same chemistry. For example, we don’t want them to jump from Next GEM to GEM-X in the middle of a project.
[Single cell] assays are sensitive to factors like cell handling, dissociation, and RNA integrity. Even small variations can lead to large downstream effects. That’s why fixation-based workflows such as Flex have been a real step forward: they stabilize samples at the point of collection, reduce variability introduced during processing, and make it much easier to reproduce results between experiments or across sites.
Many users underestimate data analysis and data interpretation. We usually address this as a potential concern, that the complexity of the analysis can be intimidating. We try to prepare them early, before data generation, by explaining what biological questions can realistically be answered and what kind of design, depth, and replicates are needed to support them.
To lower the analysis barrier, we provide structured quality-control reports and standardized data packages that can be directly explored in our online visualization browser, which is complementary to Loupe Browser. We offer training sessions in Seurat to help them interpret results and fine-tune the analysis. We also maintain close ties with internal bioinformaticians so researchers can move smoothly from raw data to biological insights without getting lost in technical complexity.
How do you help your researchers determine the right option for their single cell sequencing project?
We always start with a joint consultation to define their biological questions, and their desired resolution. What are the species, the tissue, the cell types, the number of cells that they want? Do they want just the transcripts or different modalities (CITE-seq, ATAC, multiomics, etc.)?
We try to first see what they want and then we see what the constraints are. For example, cell numbers, specific tissue dissociation considerations, will there be cell types that are notoriously difficult to handle? Do we have fixation options to postpone a lot of the biosafety level considerations? And then the conversations will always end with what's the budget that we can set aside for that.
We then map the best-fit platform: Is it droplet-based? Is it microwell-based, plate-based? Also, do we need targeted approaches versus looking at the whole transcriptome?
The consultation phase can take a while. We try to be very well prepared on that because we have a lot of hands-on experience with sorting, isolation, specific tissue digestion protocols, and we have a very large internal data repository from the core facility and from VIB for nearly 8 years worth of projects. We have quite a good sense on what to expect when people email us about a specific tissue. We can go back to our previous projects before that consultation meeting so we can know that we're prepared. We also want them to know and see that we focus on what to expect from their cell types and what the pitfalls and the problems will be.
If we have serious constraints or concerns, we will suggest a pilot experiment. Like one or two samples, technical pilots for quality control.
What features of the Flex Gene Expression assay did you feel would be most beneficial to the projects that your core regularly works on?
There are three main features that are important. The first one is the ability to fix and to biobank samples so that you have flexibility in the scheduling. This can be very transformative for multi-center studies or for multi-site operations—even our core facility has multiple sites. It's a chemistry that tolerates cryopreservation. For us, it is extremely important to make sure that sample collection is uncoupled from the library prep.
We also saw that Flex provided a very excellent data quality. I think this is something that people didn't expect, that it was almost superior quality.
And then the third advantage that led people to choose Flex for our portfolio was the lower cost because of the multiplexing strategy.
Flex provides operational flexibility and scalability at very reasonable costs, but without compromising data quality.
I think the fixation benefit is what we discovered first, and then, gradually, we realized it has super nice quality and is very consistent.
It has been 2, maybe 3 years, and we have not had any failures related to the assay. We’ve had two failures in maybe hundreds of projects, and it was sample-related.
What benefit do you find for your users in decoupling sample collection and library prep? How does the Flex assay support this process?
Flex has become more than a chemistry—it’s really an enabling workflow that extends single cell capabilities far beyond our own lab’s walls. We advertise Flex as the new way of storing samples, not just sequencing them. In our institute and ecosystem, this induced a natural evolution in how biologists think about sample preservation and readiness for future analysis. Think about how people have traditionally preserved biological material: they snap-freeze tissue, use RNAlater (a storage reagent), or fix samples for histology. Flex fits right into that same mindset, but for single cell suspensions.
In our environment, many labs already generate single cell suspensions routinely for flow cytometry or sorting. So we continuously challenge researchers: why not preserve those same suspensions in a way that keeps them ready for sequencing later on? We distribute small fixation kits and a one-page protocol.
These are homemade kits with individual reactions for each sample. We have separate “boxes” with 10 tubes already together. Each buffer from 10x Genomics is aliquoted for a one-per-sample basis. And on each tube, the protocol is also written (for example, add XX µl H2O and XX µl 37% PFA, etc). We also provide researchers with 50% glycerol (pre-aliquoted) and we sell small 37% PFA tubes alongside if needed. We actually give them everything they need! And, although this is very rare, for some labs we pass along one big 48-reaction kit.
This has become the new standard for sample archiving in our ecosystem. The samples are stable and can come back weeks or months later when the project, budget, or biological question is ready for it.
Dr. Vandamme has graciously provided access to the core’s sample fixation protocol here.
Are there any exciting projects or research findings that the core has been a part of that have used Flex that you can tell us about?
As a core, we're not focusing on a single project or a single biological question. We have hundreds of Flex-based projects that are across many tissues, across many sample types, cell types, and so on. And each brings something unique for us. The data and the biological interpretation also doesn't stay with us, so it's maybe not so easy to say that this is the breakthrough that we are focusing on.
But for us, it's very important when we have a technical breakthrough, where we have optimized something in the workflow and made things logistically feasible so that it unlocks something for our customers. When we have taken away the bottleneck, that is, for us, the moment that we have a lot of pride. It is very rewarding for us.
We take the most excitement from samples that are logistically difficult to handle and very often fragile. When these researchers are very worried that we won't be able to process them, it is quite rewarding that we are able to calm them down and reassure them that we can, even at the last minute, help them preserve a very, very precious sample and make the study very cost-effective and feasible. We give them feasibility when they don't expect it.
One area that we're really excited about is the method snPATHO-seq. We're using FFPE tissues as input for Flex, so we can actually access bioarchived material. We can access samples that were not intended for single cell sequencing, that people didn't think about doing single cell when they were harvesting and fixing these tissues. This opens up a lot of new cohorts which were previously inaccessible.
We're rolling out a lot of protocols on human and mouse samples using FFPE material as input. We had struggled a lot with pancreas samples, but now we have seen that the pancreatic samples in Flex are better than reverse transcription-based assays or single nuclei. We are also seeing that samples that have a lot of neutrophils and granulocytes and these cell types, which are quite fragile, are also being quite well preserved. So we are confident in the assay, the fixation procedure, and the GEM-X workflow used for single cell encapsulation. And in Flex now, we're using custom probes. We're using CRISPR now also on Flex.
These are the things that are very exciting at the moment and it keeps evolving. It's not just a matter of we fix and we do traditional sequencing. We try to see whether the assay can do new things, and often we manage to get it working.
Do you have any additional tips for quality control when using FFPE material with the Flex assay? How do you ensure the highest quality results?
FFPE material works surprisingly well with Flex. The first thing we do is check the age of the samples and how the samples were originally prepared. Following consistent fixation and embedding routines is often more important than any downstream optimization. We can also assess RNA quality using the DV200 score.
Several incoming samples are QC-checked in parallel for both Flex and Visium HD. If there’s any doubt, either on our side or from the researcher, we encourage doing a small pilot test first. If that pilot performs well and the project later scales up, we credit or discount that pilot cost. It’s our way of maintaining scientific integrity and protecting both our and the researcher’s reputation—because nobody benefits from processing a large batch of poor-quality samples at high cost.
You have been doing some work in your core that further reduces the price per sample of Flex. Tell us about that.
We mix and match small or low-plex kits with higher-plex kits. We mix and match so that the barcodes are actually managed separately from the GEM-X reagents. So we have a complete stock of barcodes and GEM-X reagents. I know how many reactions I can do. I know how many samples I can do. And what we offer is a flat linear pricing model, so that people can do 1, 2, 3, 4, 5, 6, however many samples. So we’re not restricted to 4 and 16. We had the flat rate pricing, and it's incremental per sample, it becomes cheaper per sample.
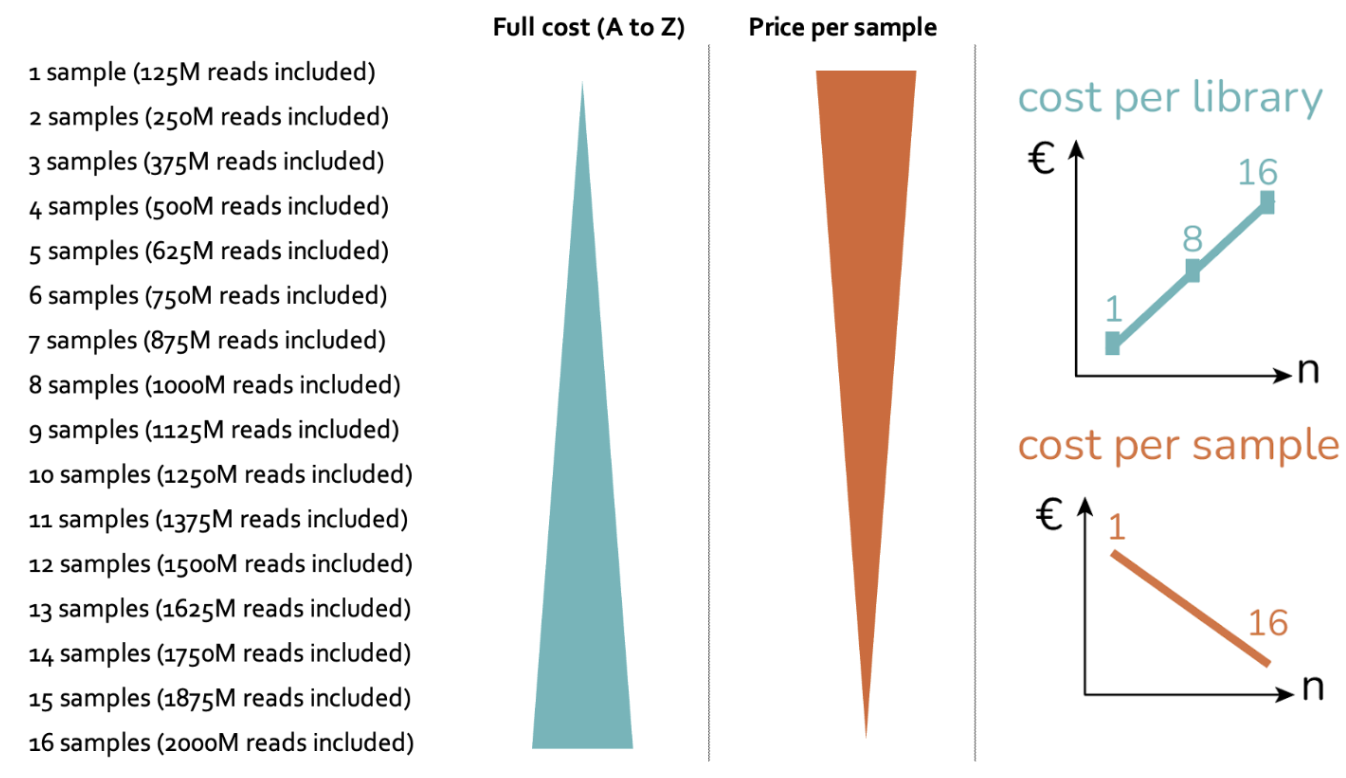
It's very nice for the researcher that they get an all-in price. They pay according to the number of samples they actually provide, with a predictable, incremental cost for sample numbers between 1 and 16, including a basic sequencing package, rather than being restricted to fixed bundle sizes like 1, 4, or 16. This gives versatility and transparent expectations for the full workflow.
Where do you see the most growth potential for single cell sequencing and/or the Flex assay? Are there any new features you’d be particularly excited to see?
If we talk about Flex, an automated, walk-away sample prep or sample processing station or a solution would help the core to scale up. There's a lot of manual work on the samples after the fixation, so that's something we are actively looking into: how to bypass by using all the types of upstream quantification methods and upstream quality control before you do the hybridization, before you do the pooling.
I think multiplexing for Flex will eventually come with 96-barcode or 384-barcode kits, enabling more scaling.
What's the growth potential for single cell sequencing, in general? Five years ago everybody was talking about spatial and multiomics, like with the RNA, ATAC, protein, methylation, and so on. I think this is still valid, although I believe the growth will not come from one breakthrough anymore. It will be from converging the things that we have already, in much more detail.
So it's convergence, using more integrated multiomics. Right now we have very limited projects that are using all these biological layers. I think the multiple layers should be combined much more, and at the level of data analysis using AI tools, to populate to existing atlases, or to populate to existing data out there.
I think Flex will play a very central role in that, enabling standardized sample biobanking and fixation for future studies. I am looking forward to seeing robust, fixation-based methods for other assays beyond gene expression and protein.
Keep exploring VIB Single Cell Core services or take a look at the Flex Gene Expression assay.
Additionally, 10x Genomics has recently released plate-based multiplexing for the Flex assay, offering modular kit options with flexible batching and independently sold core reagents to fit your specific experimental design, which can be beneficial to many researchers and core labs.

New product! Explore Flex, reimagined
The new Flex assay delivers plate-based multiplexing capabilities, allowing researchers to pool up to 384 samples in one experiment for incredible scale and cost savings. Plus, modular kit options enable flexible batching and no wasted reagents.
This interview has been edited for length and clarity. We'd like to thank Dr. Vandamme for his contributions to this blog article!
About the author:

