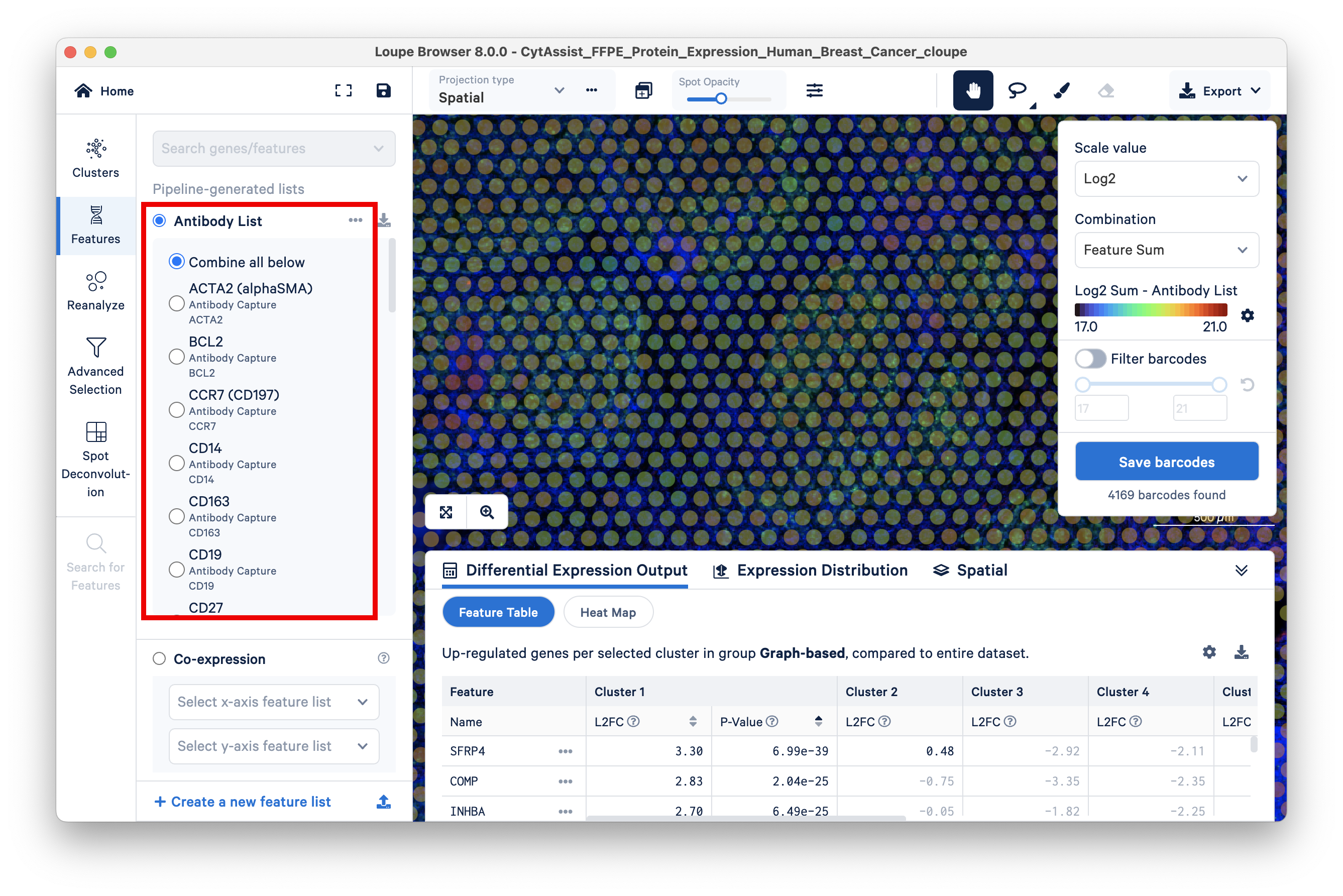
Loupe Browser v6.5 introduced the ability to filter barcodes based on the detected level of specific antibodies included in your Visium CytAssist Spatial Gene and Protein Expression workflow. Additionally, Loupe Browser's feature thresholding functionality helps further filter these barcodes based on their level of antibody (protein) expression. Antibody expression-based filtering and thresholding can help better identify and classify cell types per your criteria of interest.
In this tutorial, we will use a Visium human breast cancer dataset with Loupe Browser v8.0 to demonstrate how you can begin analyzing protein expression data.
- Use protein expression to identify and classify cell types, especially where mRNA markers have low expression.
Begin by setting the Spot Opacity slider at zero to visualize the image without overlaid spots. You may also wish to auto-adjust the brightness and contrast.
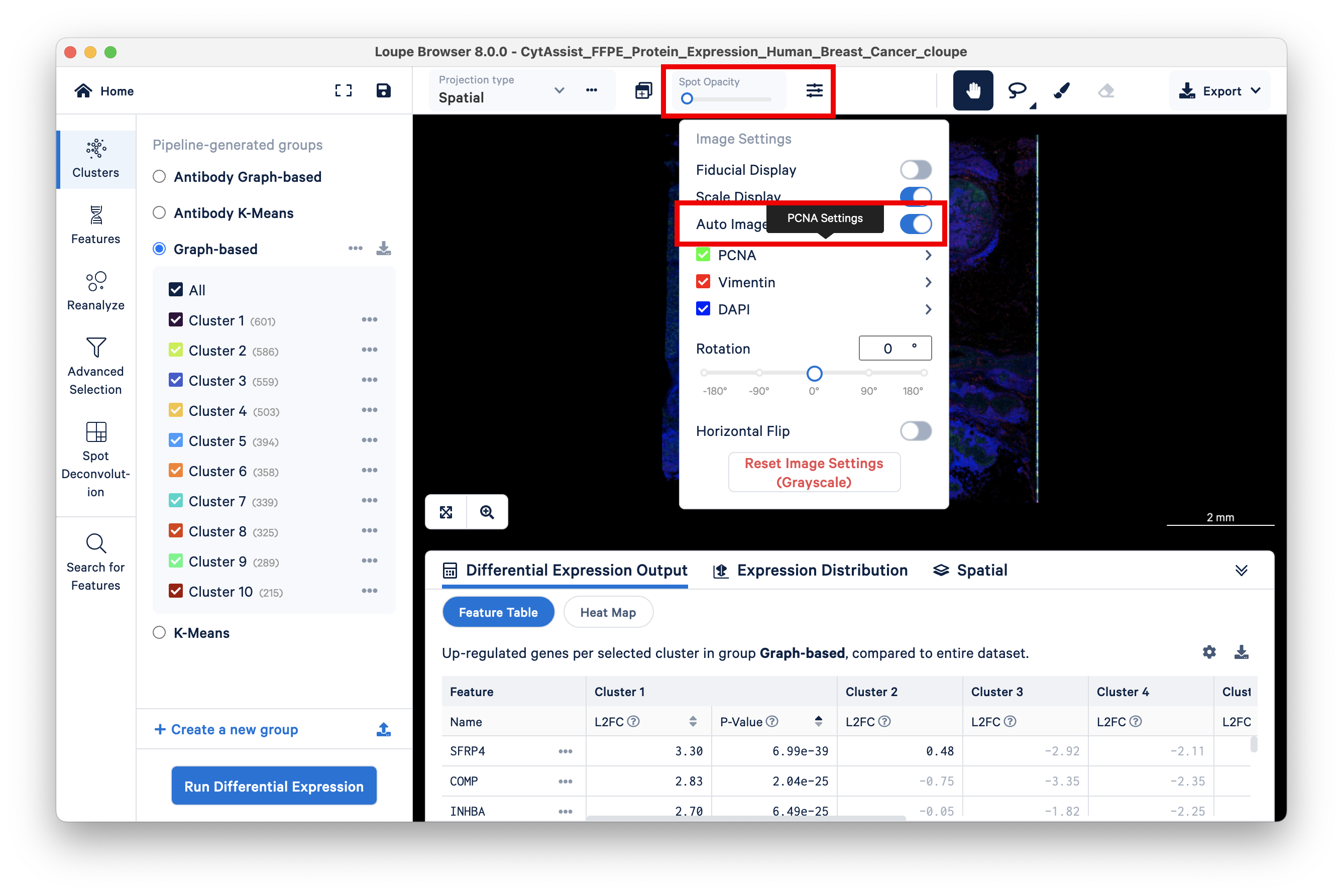
Green cells are stained with PCNA, an antibody stain that labels rapidly proliferating (potentially cancerous) cells. Areas on the tissue slice with high expression of PCNA have a larger number of green cells, as seen by setting the Spot Opacity scale to zero.
Loupe Browser allows you to search for PCNA-expressing barcodes and color code them by the number of UMI counts (heat maps). To do this, click on Features and search for PCNA. One option for the Gene feature and one for the Antibody Capture (Protein) feature should appear.
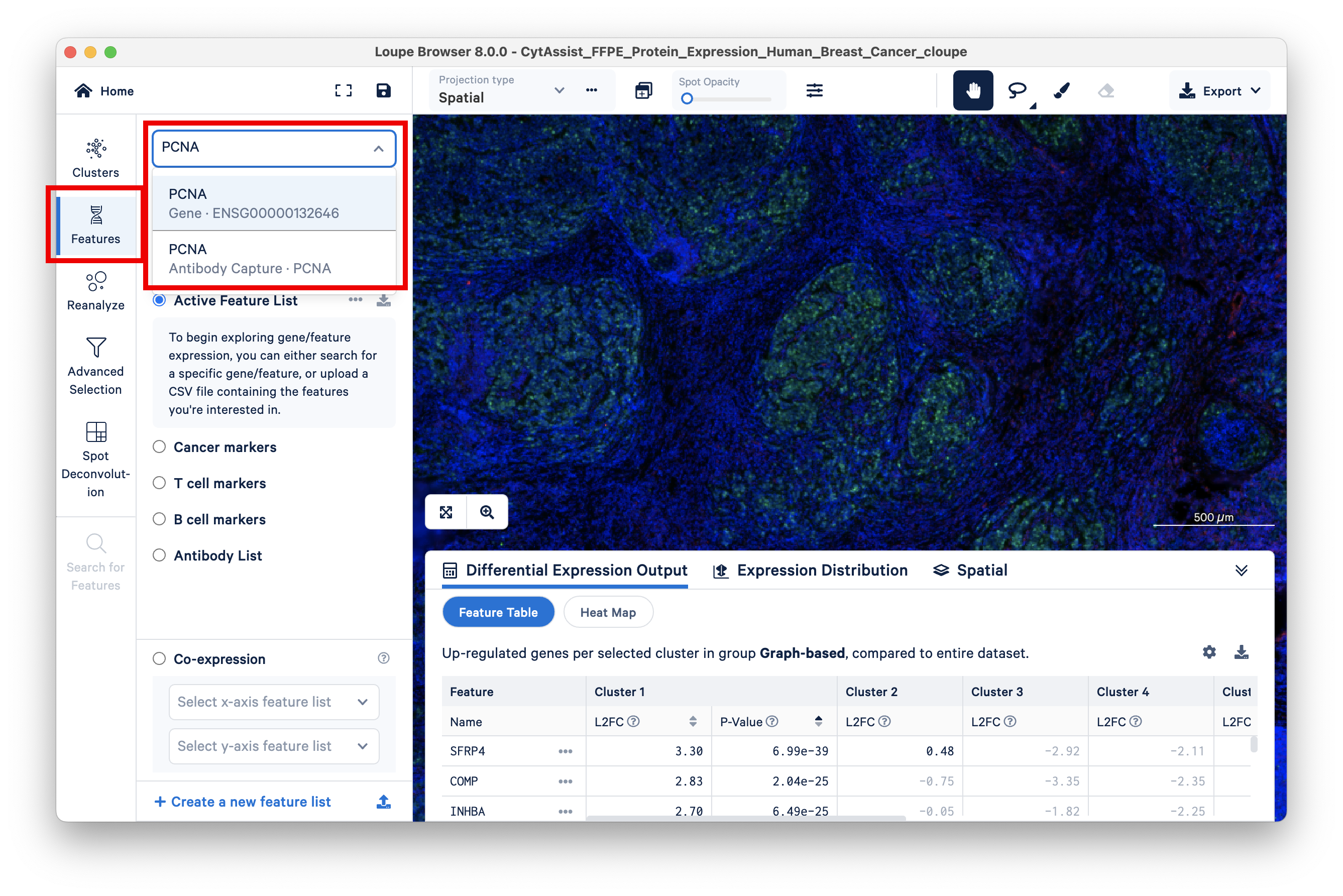
First, select the Gene feature.
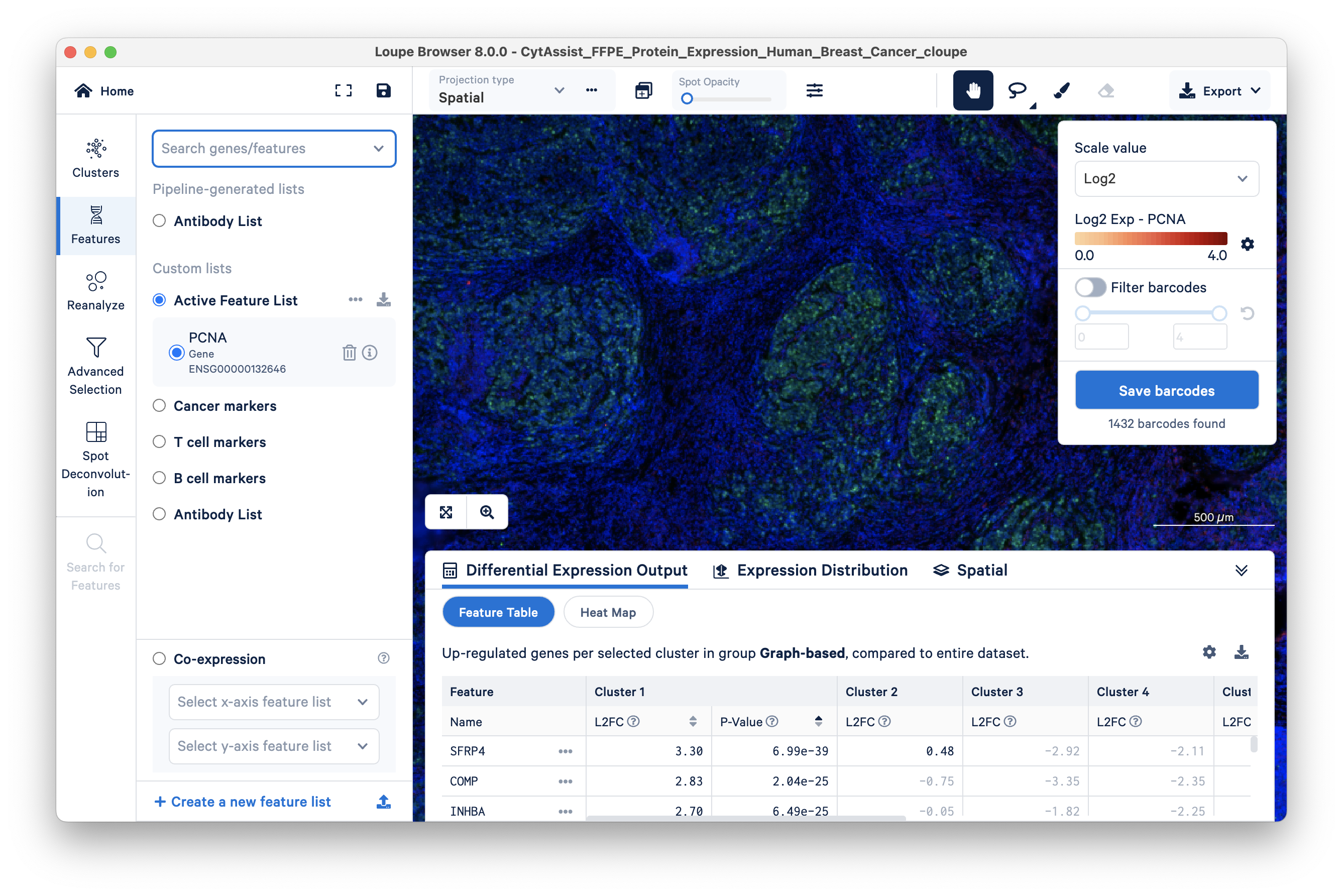
Gradually increase spot opacity using the sliding scale, and change the scale from Log2 to Linear. By sliding the Spot Opacity scale from left to right, find the appropriate opacity to visualize both the spots and the stained tissue in the background.
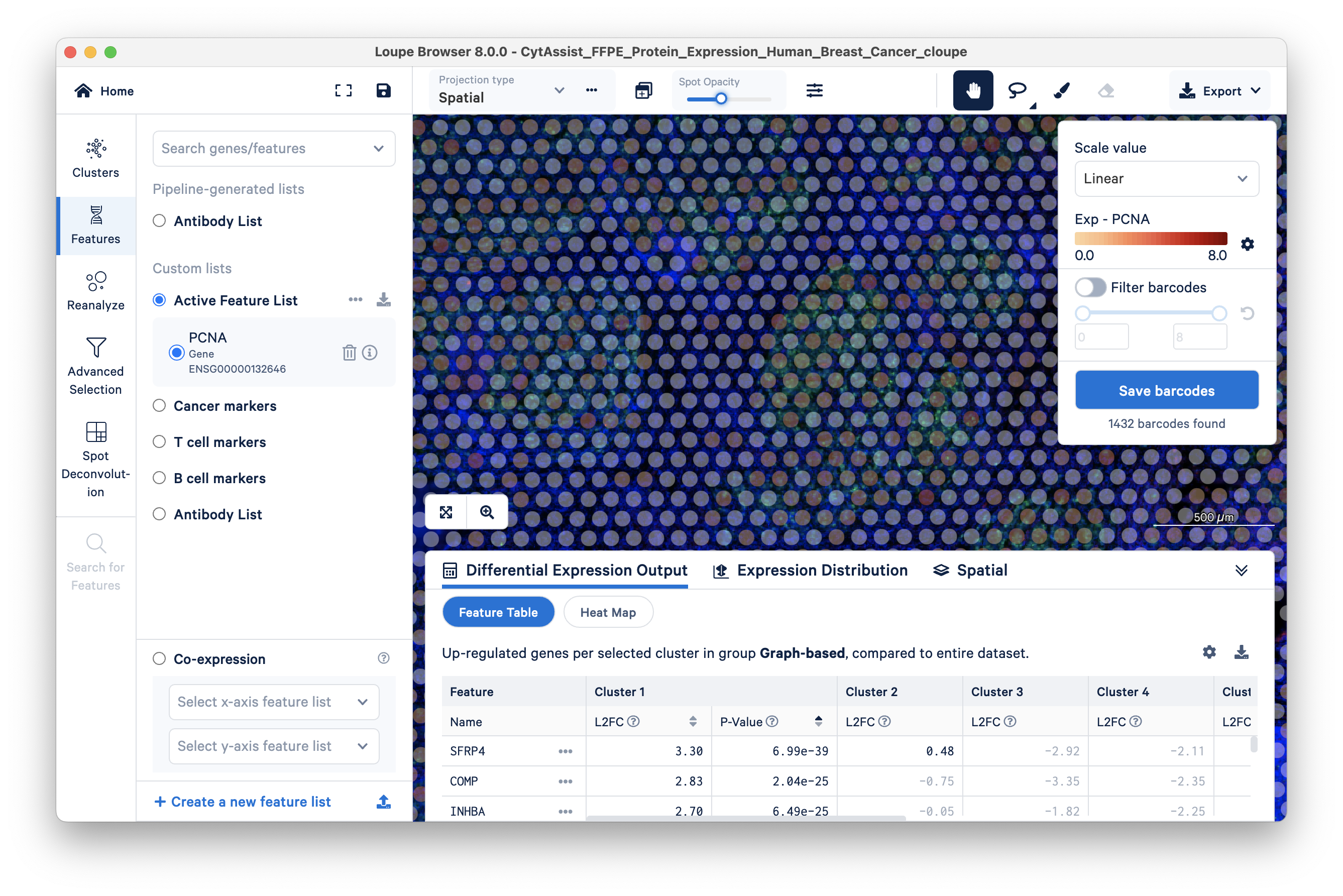
You can see that some areas with green cells have transparent barcodes (spots). This poor sensitivity is due to low mRNA expression; there is insufficient mRNA for the RTL probe-based capture, resulting in dropouts.
To check the performance of the PCNA antibody (versus the PCNA gene), use the 'Search for a feature...' box again to search for PCNA. This time select the Antibody Capture feature.
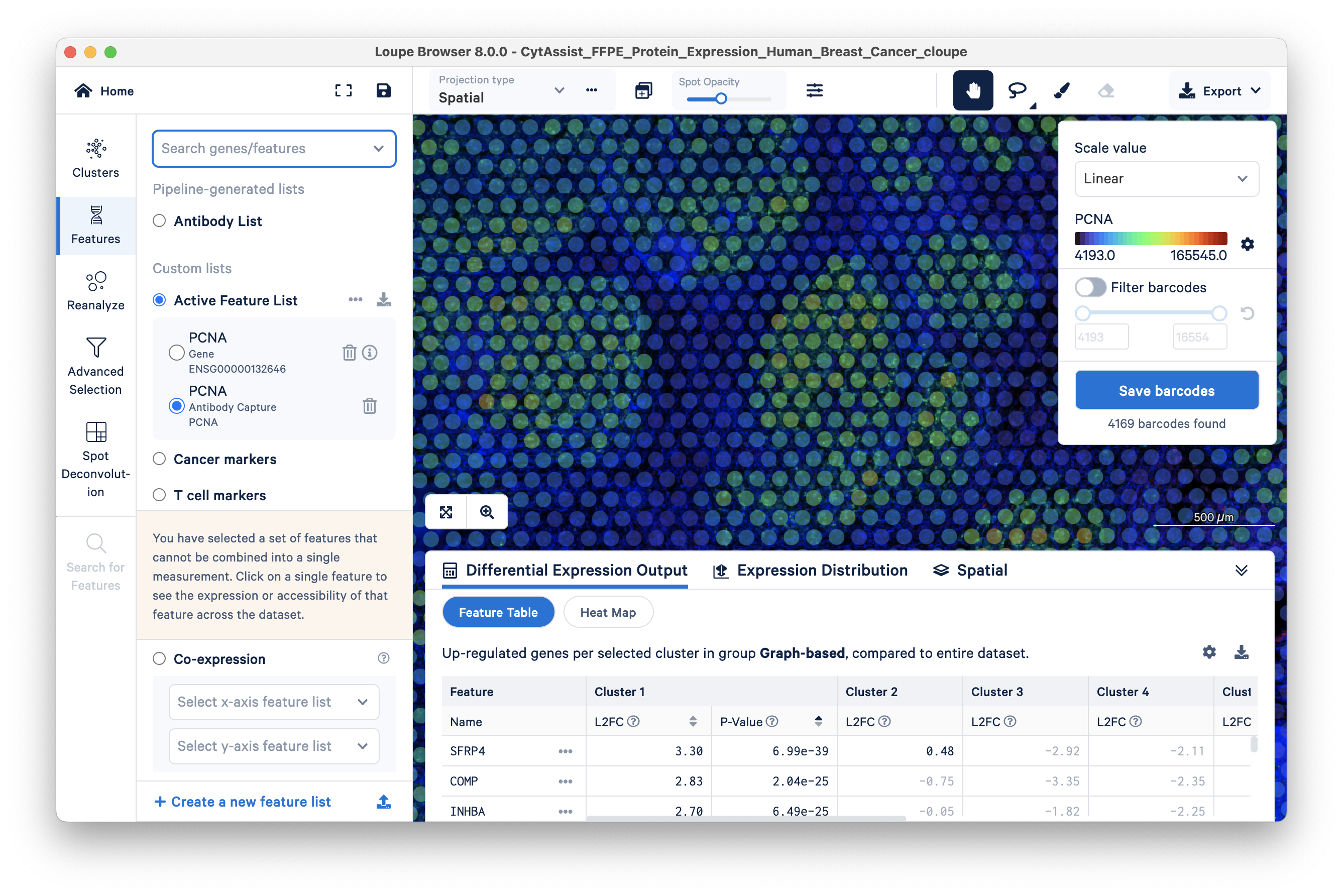
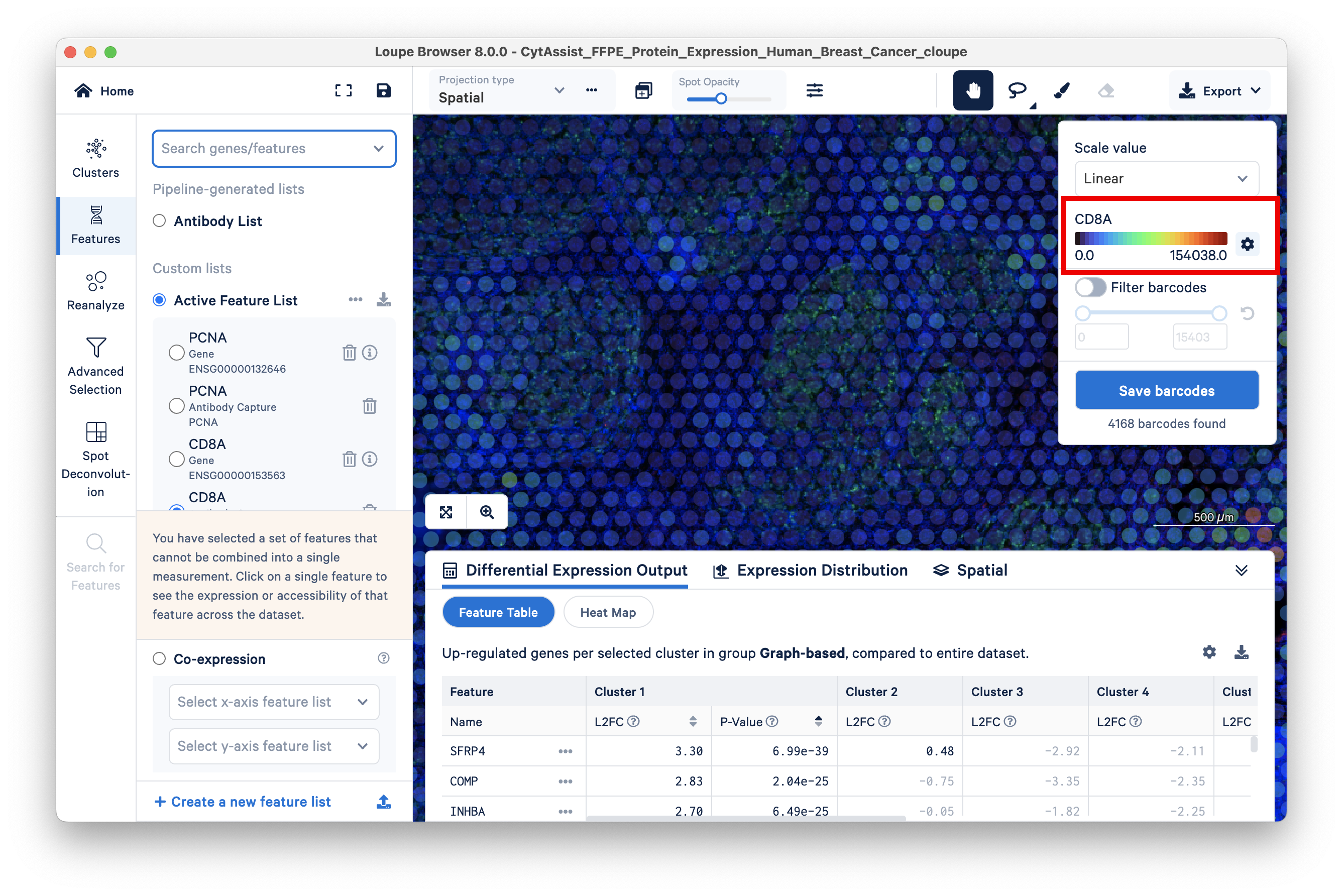
The Antibody Capture feature is generally more sensitive than the Gene feature. Given this greater sensitivity, protein expression data can be particularly helpful for classifying cells that have low RNA expression, e.g., immune T and B cells. To demonstrate this, a comparison of the expression pattern of T cell marker CD8A gene feature vs antibody feature is shown below.
The gene feature has a maximum UMI count of 4:
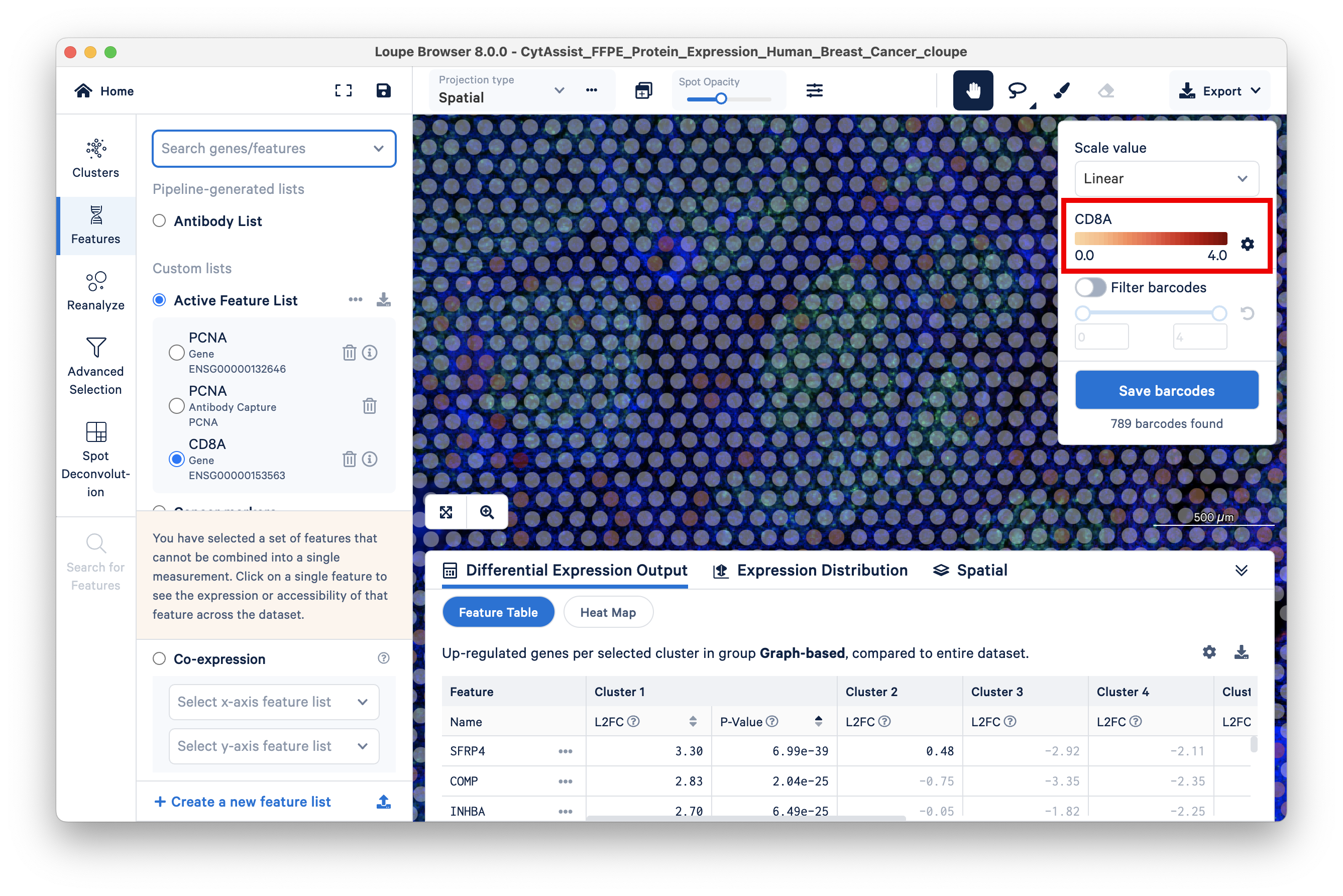
The corresponding antibody feature has a maximum UMI count of 154K:
To view the list of antibodies included in the experiment, the Features view has the option to display the Antibody List.