- The Clonotype Distribution plot now supports coloring clonotypes by sample or metadata provided in the Cell Ranger
aggrinput CSV, offering more customizable visualizations. For details, refer to the Comparing Multiple Samples tutorial. - Exported sequences for selected clonotypes now include their associated Sample ID(s), improving traceability and facilitating downstream data analysis.
- The honeycomb plot can now be exported as a CSV file, containing the X and Y coordinates for all barcodes, allowing for detailed examination and further analysis.
- Clonotype IDs are now displayed in the clonotype filter list, enabling easier tracking of clonotypes across both the web summary and output files.
- Fixed a bug that prevented imported Loupe files from being removed from the workspace.
- Fixed a bug that caused inconsistencies in clonotype numbering across pipeline outputs and the Loupe (VDJ) Browser.
- Introduces ability to visualize (Comparison tab) BEAM (Antigen Capture) libraries aggregated with
cellranger aggr.
- Bug fix to display the correct consensus sequence under the “Align Contigs to Consensus and Reference” option.
- New and improved user interface that allows for rapid filtering and exploration of V(D)J datasets!
- Supports data analysis for Antigen Capture (BEAM) with new visualization and filtering tools that enable users to rapidly explore the relationships between clonotypes and antigens.
- Immune cell clonotypes can now be visualized through the Clonotype Distribution plot.
- Updated Sequences view for improved visualization of paired BCR or TCR receptor chains along with receptor nucleotide sequences for exact subclonotypes within a clonotype and their associated antigen specificity score.
- Supports changes to Cell Ranger v5.0, including V(D)J
aggrand the new clonotype grouping algorithm. - Required upgrade for processing data generated by the Cell Ranger v5.0 pipeline. Due to extensive changes to the underlying infrastructure,
.vloupefiles generated by Cell Ranger 4.0 and earlier Cell Ranger versions cannot be read into Loupe V(D)J Browser 4.0 or Loupe Browser 5.0.
Comparison Mode:
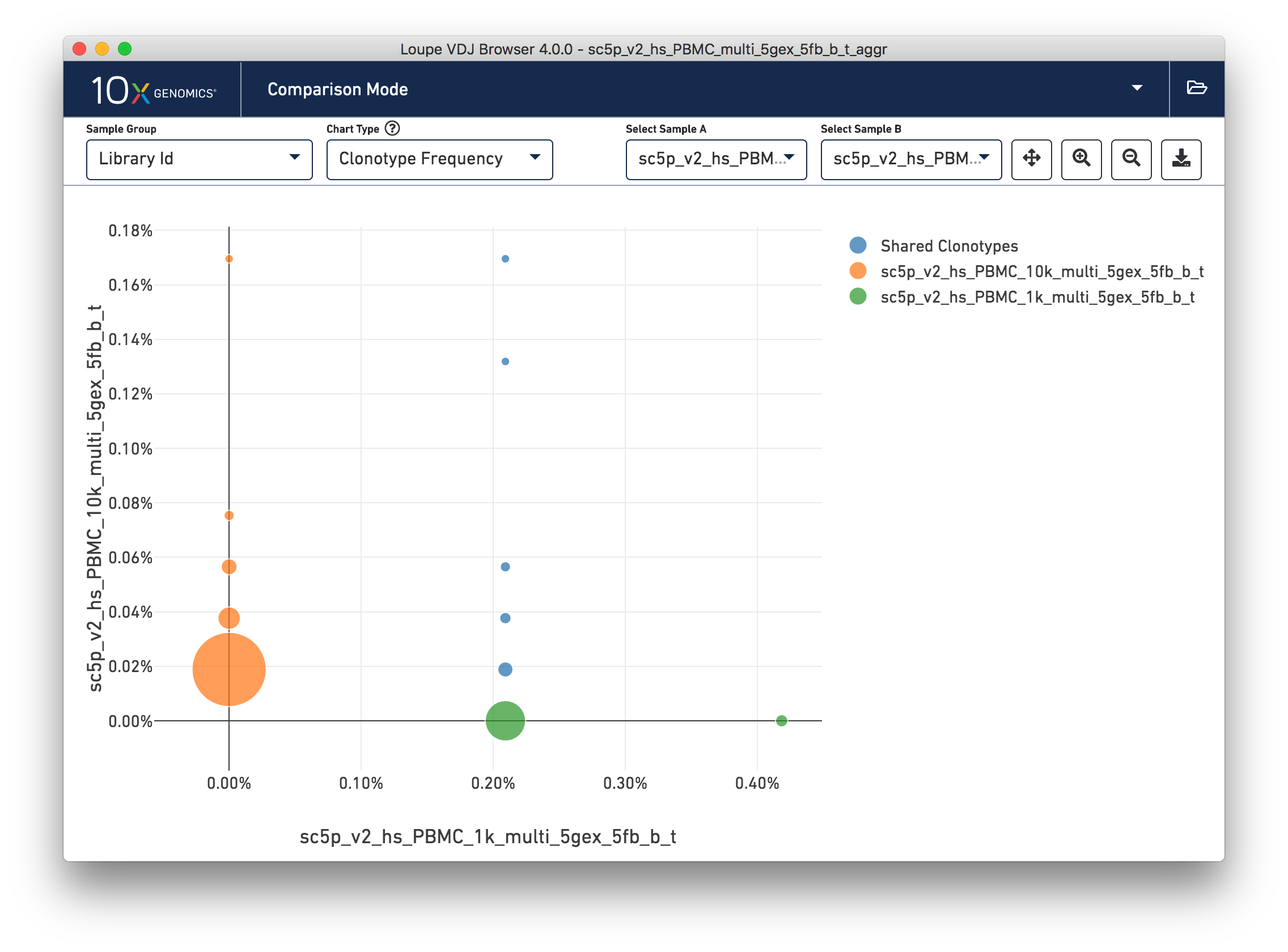
Loupe V(D)J Browser 4.0 now allows for comparison of samples in files generated by the V(D)J aggr pipeline. Data can be divided into samples based on the columns in the aggregation CSV provided to the aggr pipeline, such as donor, origin, library ID, and any custom columns. A sample group dropdown has been added to all comparison mode charts, allowing for quick navigation.
For .vloupe files with a large number of samples, a new selection option will appear to enable displaying a subset of samples in a single view.
The clonotype frequency view is no longer accessible via a button in the sample table, but can still be found in the chart type dropdown. Selectors have now been added to the clonotype frequency chart to allow for quick switching between samples.
Clonotype Table Filtering:
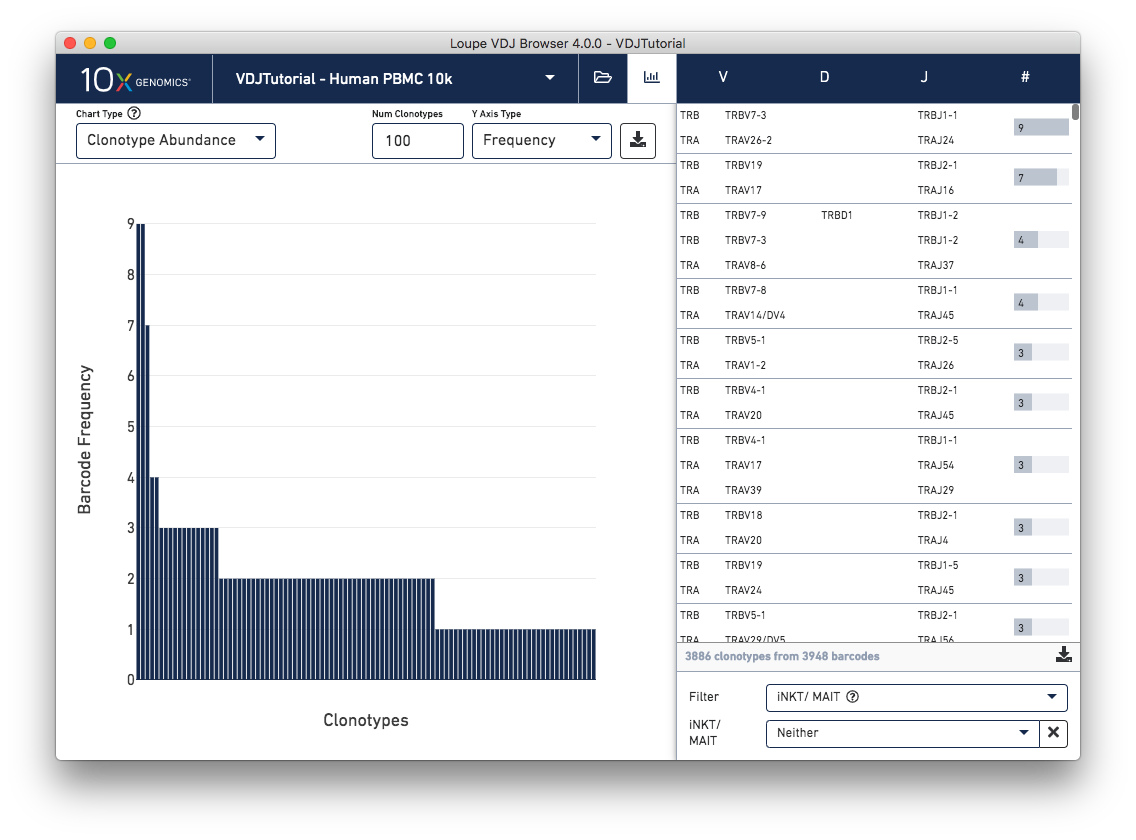
Identify potential invariant natural killer T (iNKT) cells and mucosal-associated invariant T (MAIT) cells with a new clonotype table filter. The iNKT/MAIT evidence filter can be used to view clonotypes that contain any cells with supporting evidence in any of the TRA genes, TRA junctions, TRB genes, or TRB junctions of being an iNKT or MAIT cell. This filter is only visible for T cell datasets.
There are a number of other powerful filtering options added in this release:
- The clonotype table can now be filtered by sample for
.vloupefiles generated byaggr. - Clonotypes in B cell datasets can be filtered by light or heavy chain isotype.
- Graph-based clusters generated by the new reclustering feature in Loupe Browser v5.0 can now be used to filter the clonotype table as part of the existing cluster filter.
Chain View Updates:
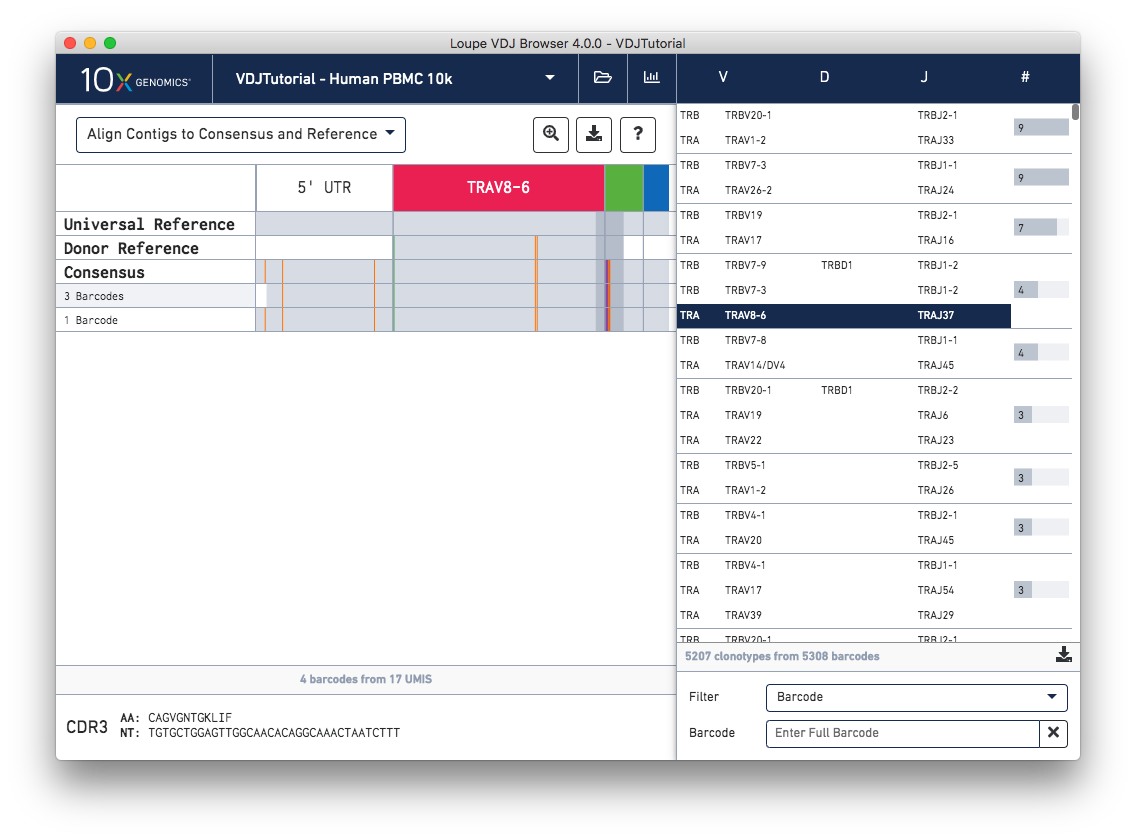
It is now easier than ever to gain insights from the chain view. Rows in the chain view now represent exact subclonotypes instead of individual cells, meaning cells with identical immune receptor nucleotide sequences are now collapsed into a single row in the table. A Download Barcode List option is available when hovering over an exact subclonotype to see the full list of barcodes included in the row. Additionally, a donor reference is now visible between the universal reference and consensus. The donor reference only spans the length of the V gene.
Additional Features:
- Improved performance across the application on datasets with a high number of cells.
- Refreshed appearance for several features.
Removed Feature:
- The IGV plugin view has been removed from Loupe V(D)J Browser v4.0.
- Loupe V(D)J Browser v3.x is a required upgrade for processing data generated by the Cell Ranger v3.0 pipeline.
- Same UI features as Loupe V(D)J Browser v2.1.0.
- Import
.cloupefiles generated by Cell Ranger v3.0.
- Better support for allele-aware references.
- New B cell paired and unpaired isotype summary charts.
- Convert all proportions to percentage.
- Clarify help clonotype list, comparison, and bug report text.
- Renames Multi-Sample Comparison to Comparison Mode
- Fixes Sample Overlap single-chain counting bug, simplifies similarity metric options
- Prevents click event from comparison mode selector from propagating to the chart underneath and hiding a sample
- Comparison Mode documentation update
B Cell Support:
- The Chromium™ Single Cell V(D)J Solution now offers comprehensive, scalable profiling of both 5′ gene expression transcripts and full-length, paired T Cell Receptor and B cell immunoglobulin transcripts. Loupe V(D)J Browser has added a few additional features especially designed to help with analyzing B Cell samples.
- In chain view, indicate Isotype Switching alongside the barcode labels.
- All existing features in Loupe V(D)J browser are compatible with B cells.
Comparison Mode:
Loupe V(D)J Browser v2.0 allows you to load multiple samples into the same workspace and includes powerful plots, metrics, and statistics to compare multiple samples. You can also quickly switch between multiple samples a single workspace. New features include:
- Clonotype Comparison - Compare the relative abundance of clonotypes between two samples and find the clonotypes that have the greatest difference
- Sample Overlap - Determine sample similarity between paired clonotypes or unpaired chains
- Top Clonotype Overlap - Determine which top clonotypes are shared between multiple samples
- Clonotype Frequency - Plot and compare the clonotype abundances between two samples.
- Sample Table - Quickly compare high level metrics between multiple samples
- Gene Usage - Plot the gene usage of the V, D, J, and C genes accross multiple samples.
You can also export any of these charts to an image or the underlying data to a CSV.
Integrated V(D)J Clonotype + Gene Expression Analysis:
When using the combined 5′ Gene Expression and T-Cell and B cell enrichment kits, it is now possible to measure whole transcriptome gene expression and V(D)J repertoire diversity from the same sample. To facilitate this analysis, Loupe V(D)J Browser 2.0 allows you to combine the V(D)J and gene expression data with a few clicks.
To find out more about how to conduct an integrated experiment, consult the Single Cell V(D)J + Gene Expression page. For a complete Loupe how-to including some example data, go to Integrated V(D)J and Gene Expression Analysis in Loupe V(D)J Browser.
The complete list of gene expression features in Loupe V(D)J Browser include:
- Import
.cloupegene expression files into a single.vloupeworkspace Compare clonotypes identified in gene expression clusters within one or between two samples and find clonotypes that have the greatest difference - Filter the clonotype list for clonotypes that only exist within one gene expression cluster
- On import, detect whether a V(D)J dataset is associated with the loaded gene expression data
Additional Features:
- Summary Charts
- Added new CDR3 bar graph
- Added D Gene and C Gene Usage Charts
- Export all summary charts as SVG
- Export summary chart data as CSV
- View help text for each chart by hovering over the question mark
- Export Clonotype List as clonotypes or single chains
- Support for soft-clipped alignments generated from Cell Ranger
- Added color legend for chain view variants
- Support native macOS window switching
- Augment all inputs by allowing the select all hotkey
- Fixes issue that may block file opening and bug reporting when an HTTP proxy is active on a user's system.
- Corrects title of tutorial dataset.
- Initial release.