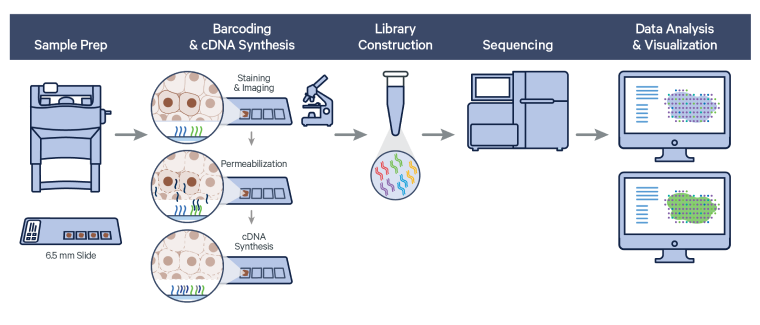
The Visium Spatial Gene Expression workflow measures RNA levels in intact, fresh frozen (FF) tissue sections, by capturing mRNA using spatially barcoded oligonucleotides attached to the Visium slide. The workflow begins by first determining the optimal permeabilization time for the tissue. The tissue is stained, and imaged followed by cell permeabilization to release mRNA which binds to the poly(dT) region of spatially barcoded oligonucleotides present on the slide surface. Libraries are generated from the barcoded cDNA of a captured transcript and are sequenced.
Visium Spatial Gene Expression relies on Space Ranger analysis software which uses STAR aligner to perform splicing-aware alignment of transcript reads to the genome. Space Ranger uses the transcript annotation GTF file to identify exonic reads, which when compatible with a single gene annotation, are considered for UMI counting.
- 1
Download and setup Space Ranger
Space Ranger is delivered as a single, self-contained tar file that can be unpacked anywhere on your system. It bundles all of its required software dependencies, which are pre-compiled to run on a wide range of Linux distributions.
- 2
Input FASTQ files and Image
Before you begin your Space Ranger run, you will need to prepare the input files. These files will vary depending on the parameters of your experiment, the assay chosen, and which Space Ranger pipeline you are using.
- 3
Run pipelines
Learn how to set up, run, and explore outputs using spaceranger count for FF.
- 4
View and download outputs
Understand output structure and files summary.