A pathologist’s perspective: Advantages of using FFPE tissues for single cell analysis
*Impact at a glance: A pathologist shares how 10x technology provides a deep, cellular-level view of common preserved tissue samples, connecting decades of patient records directly to what’s happening in their cells. This means doctors and scientists can learn more from existing samples, speeding up discoveries and improving care for future patients.*
How do pathologists approach critical questions of disease diagnosis? What tools help them move closer to a detailed understanding of the cellular processes that lead to disease? Traditionally, histological methods have driven diagnosis, but, in recent years, highly sensitive molecular assays—including single cell analysis—have begun to provide avenues for more in-depth profiling of tissue complexity. With these complementary methods, the field gains more potential to move toward personalized medicine.
One commonly held requirement for many single cell gene expression profiling methods is the use of fresh samples. For a pathologist, this can present some logistical challenges because your study can only begin when you start collecting tissue, a workflow that requires careful planning and coordination with collaboration partners in the clinic. Furthermore, prompt action must be taken to either snap-freeze the samples or move to dissociation and processing on the same day.
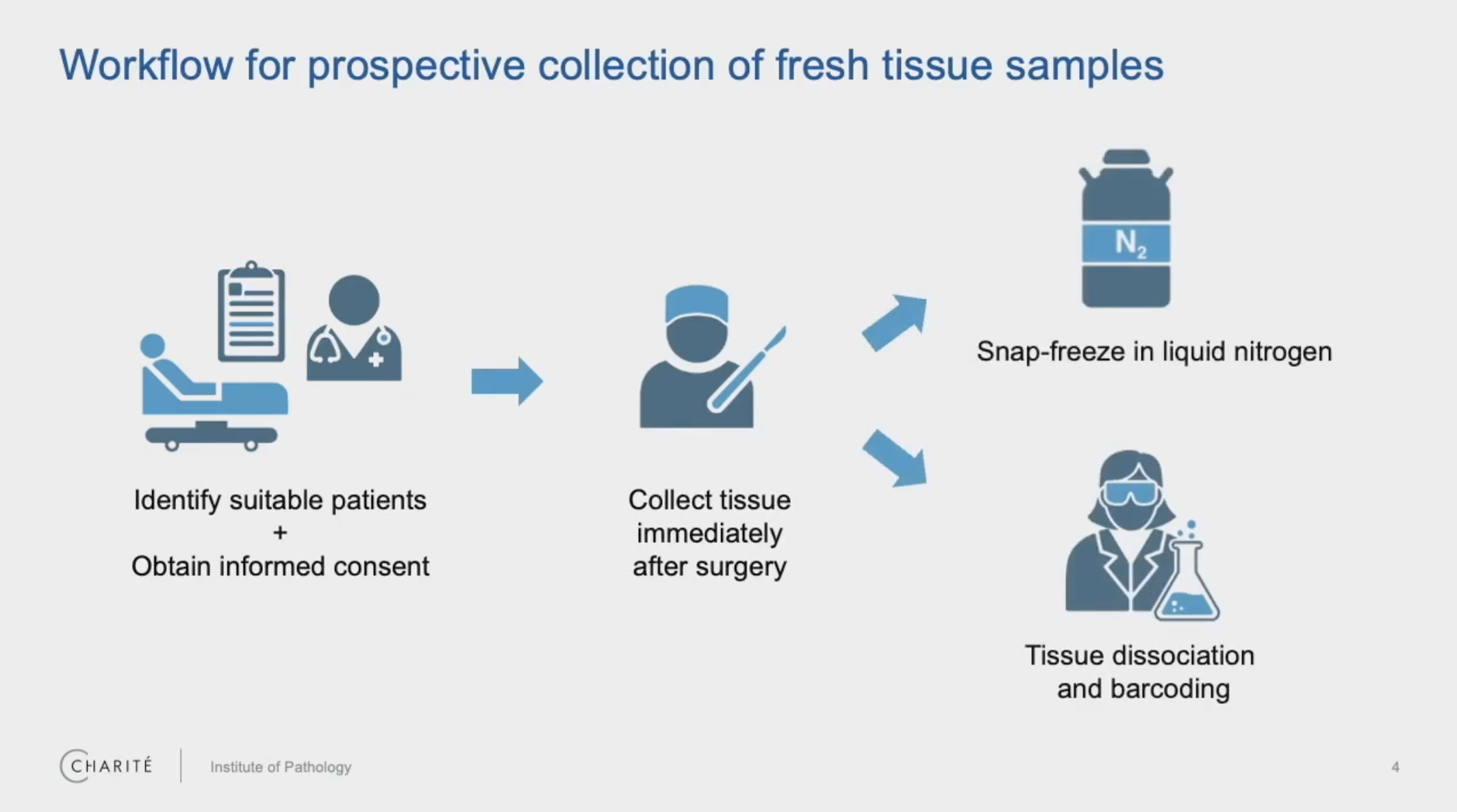
In addition to the time and effort involved here, this prospective sampling can be limiting in terms of available sample types. Information that could determine the specific direction or scope of your research—like tumor subtype or driving mutations for cancer studies—may not be available for days or weeks after surgery.
Bringing single cell for FFPE samples to the field of pathology
Dr. Philip Bischoff, a resident pathologist and clinical scientist at Charité University Hospital in Berlin, studies the intricacies of lung tumors, with specific focus on the tumor microenvironment. He has been exploring how the use of FFPE tissue can transform the impact of single cell analysis in the field of pathology, enabling the study of retrospective samples. Bringing FFPE samples into the picture means the ability to address important clinical questions by selecting patient samples based on defined criteria, including disease status, specific mutations, treatment details, and patient responses.
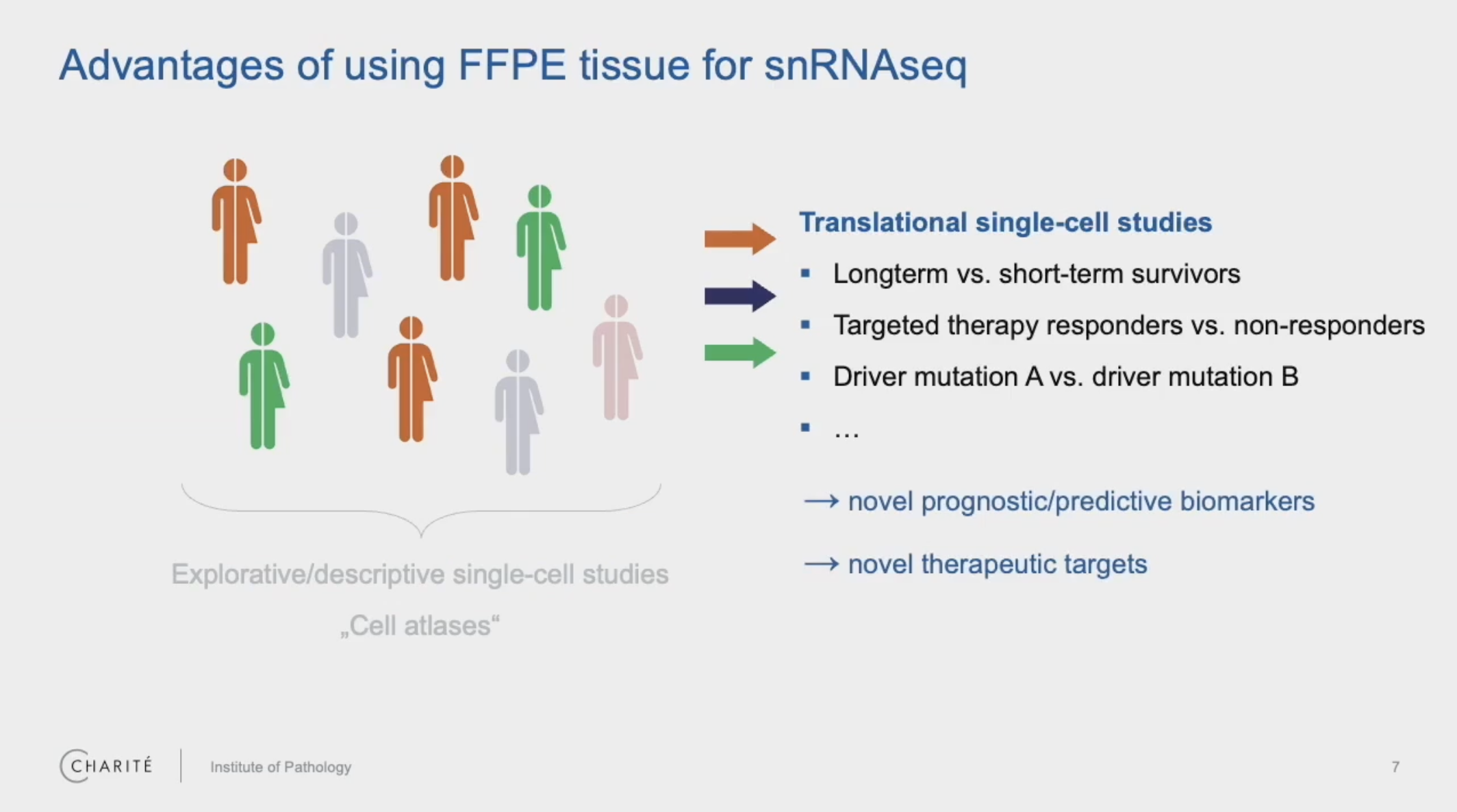
In a recent webinar, we spoke with Dr. Bischoff, who shared his experiences in analyzing these sample types with Chromium Single Cell Gene Expression Flex. His team systematically compared fresh and snap frozen tissues of three non-small cell lung cancers and corresponding FFPE tissue blocks from exactly the same tumors from their archive. The resulting scRNA-seq data allowed them to examine cell-type quantities and gene expression patterns between the sample types and also delve into correlations between gene signatures and different patient tumor morphologies.
Watch the full, on-demand webinar now to learn more about:
- Benchmarking data between sample types and factors that influence data quality
- Observed relationships between transcriptomic profiles and more aggressive tumors
- Experimental considerations and protocol optimization
- How single cell can help researchers move forward in their translational studies
Keep reading for a look at some of the questions Dr. Bischoff answered during the live Q&A session, plus some additional insights from Dr. Angela Churchill, the Product Marketing Manager of Single Cell Applications at 10x Genomics.
Advantages of single cell RNA sequencing on FFPE samples
Are there any tissues that give low-quality results for single cell dissociation from fresh samples that give better or give higher-quality data when using the Flex reagents?
Churchill: Yes. When you're starting with fresh tissue, you have a couple of different options in terms of sample preparation. Researchers could dissociate into single cells, they could isolate the nuclei, or they could use a Chop/Fix method, as we call it.
When it comes to samples that might be challenging to dissociate or might have lower-quality RNA, that's when Flex may perform particularly well, just given that probe-based detection method and the fact that it only requires that 15-nucleotide footprint with the probe design.
Were any cell types discovered in the FFPE tissue that were missed in the fresh tissue (maybe damaged in processing)?
Bischoff: The cells that are suffering the most from the mechanical dissociation of fresh tissue are epithelial cells because epithelial cells are really used to growing in a tissue. And when they are dissociated, they go into apoptosis, and we see higher scores of cell stress signatures in the fresh tissue, which you do not see in the FFPE.
Instead in the FFPE tissue, we see more fibroblasts, which is also interesting because fibroblasts are interesting cells for a lot of cancer researchers. But it's difficult to dissociate fibroblasts from fresh tissue because they have a lot of extracellular matrix around them, collagen, and it's difficult to dissociate this. It's easier to obtain these cells from FFPE tissue.
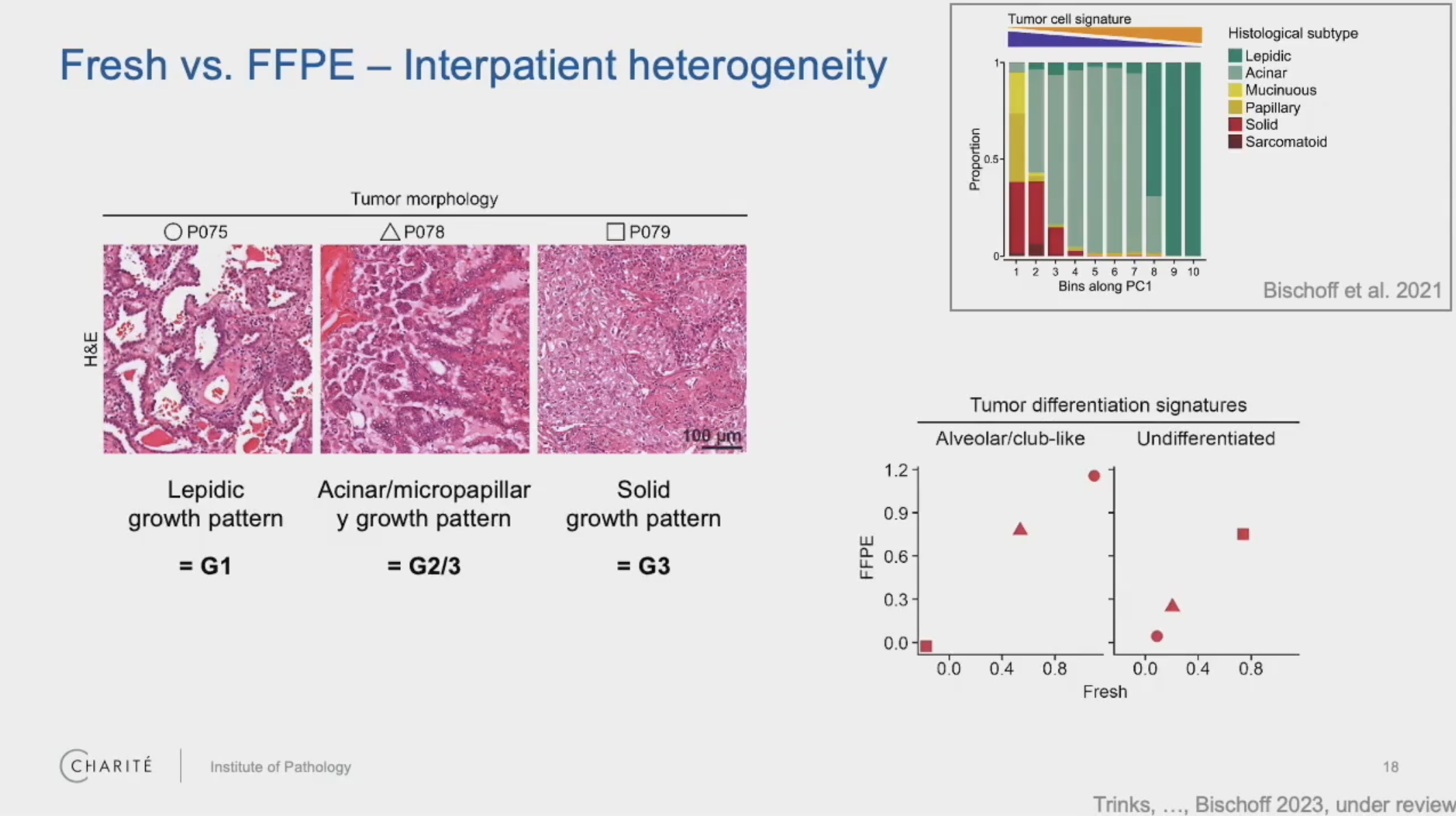
For your study [of FFPE human lung tumors], could you explain the relationship between the tumor morphology and the [single cell] gene [expression] signatures?
Bischoff: We found that in the tumors showing more differentiated morphology, these tumors are characterized by genes which are also found in normal lung epithelial cells. So this gene signature reflects that these tumors are more closely related to normal lung epithelial cells.
Then, in the poorly differentiated tumors, these genes are only very lowly expressed. And instead, we find gene signatures which are reflecting higher signaling strength of EGFR signaling, for example, and TGF-beta signaling. But the gene signatures of normal epithelial cells are lost in these poorly differentiated tumors.
Single cell workflow considerations for FFPE
Regarding the dissociation steps, did you use an Octo Dissociator or a pestle-based [method]?
Bischoff: We used an Octo Dissociator, but we also used the pestle-based protocol for smaller tissue samples. And both of them worked for us.
Could you please elaborate more on your experience with dissociation of different tissue types? Particularly, are there different dissociation settings based on tissue type and the thickness of the FFPE slice? (Something that you would program on a gentleMACS instrument?)
Bischoff: So our experience is still limited. We have not tested a lot of tissue, but what I can say is that, if you have tissue which has a high amount of collagen matrix—for example, breast tissue has a lot of fibrous tissue in it—it's more difficult to dissociate these tissues. And what helped us was just to prolong the dissociation time. So we just ran the program on the Octo Dissociator twice. We also used a program that is a bit more harsh, which rotates at a higher velocity. So, yeah, we used this to optimize our dissociation protocol for breast tissue. And this might also be applicable to other tissue types.
Can you elaborate on the possible stops in the workflow from FFPE block to finished sequencing?
Churchill: Prior to partitioning into single cells in the instrument, there's basically two optional stopping points. One is after fixation—so, [in this case,] the actual FFPE block. Then there's another optional stopping point after the probe hybridization step.
Bischoff: Yeah. And as I showed you, we, in particular, used the second stopping point for adding an additional FACS sorting step, which was very helpful for us in counting the cells and also removing debris.
Do you include DAPI for FACS sorting since it is an FFPE sample?
Bischoff: Yeah. We used DAPI staining for the nuclei, and it didn't impair our analysis.
Determining factors for data quality from FFPE blocks
How does the age of FFPE blocks affect the results, and how could one go about quality control in those older FFPE blocks?
Bischoff: This is a very interesting question, and I wish I [had] a definitive answer for this because this is what we are asking ourselves. I think the age of FFPE blocks might not be so important because we found that in very young FFPE blocks (which are just a couple of months old) and also in older FFPE blocks (which are two or three years old), we get the same quality.
I think it might be more important how well the tissue has been formalin fixed in the first place. So, if you have tissue which is very poorly formalin fixed, then even a very young FFPE block will give you poor RNA quality.
And the second part of the question, how can you make sure that you have good RNA quality in your FFPE tissue? To be honest, we didn't check for RNA quality before we started our single cell analysis. But I think even when you have a bit more degraded RNA, because this is a probe-based approach, you can still get quite good results.
I absolutely agree in saying that immediate fixation is critical for FFPE sample quality. What do you think about fixation time? In your experience, are longer fixation times better?
Bischoff: Longer is not always better. We did not test it in our lab, but a colleague who's also working on this told me that samples which have been formalin fixed [over a very long period of time]—a couple of days—might have poorer RNA quality compared to samples that have only been fixed for like 24 hours. So, very long formalin fixation might be a negative factor.
Do you have any experience with FFPE tissue including necrotic areas? I myself experienced a lower number of transcripts in the resulting data.
Bischoff: Yeah. This sounds plausible to me that RNA amount and quality is very poor in necrotic areas. We didn't analyze any necrotic tumors. And I would rather exclude necrotic areas from analysis because I would expect low RNA quality. And this is also the advantage of FFPE tissue—that you can have, really, a close look at your tissue and choose high-quality tissue and exclude low-quality tissue.
FFPE sample size and stability
How stable are FFPE samples after being taken from the block, cores versus curls,* specifically?
Bischoff: We have not tested this systematically. But I would guess that cores might be a bit more stable because it has less surface [area] and less surface contact to the air. That might impair the quality in curls, but this is only guessing; we have not tested this.
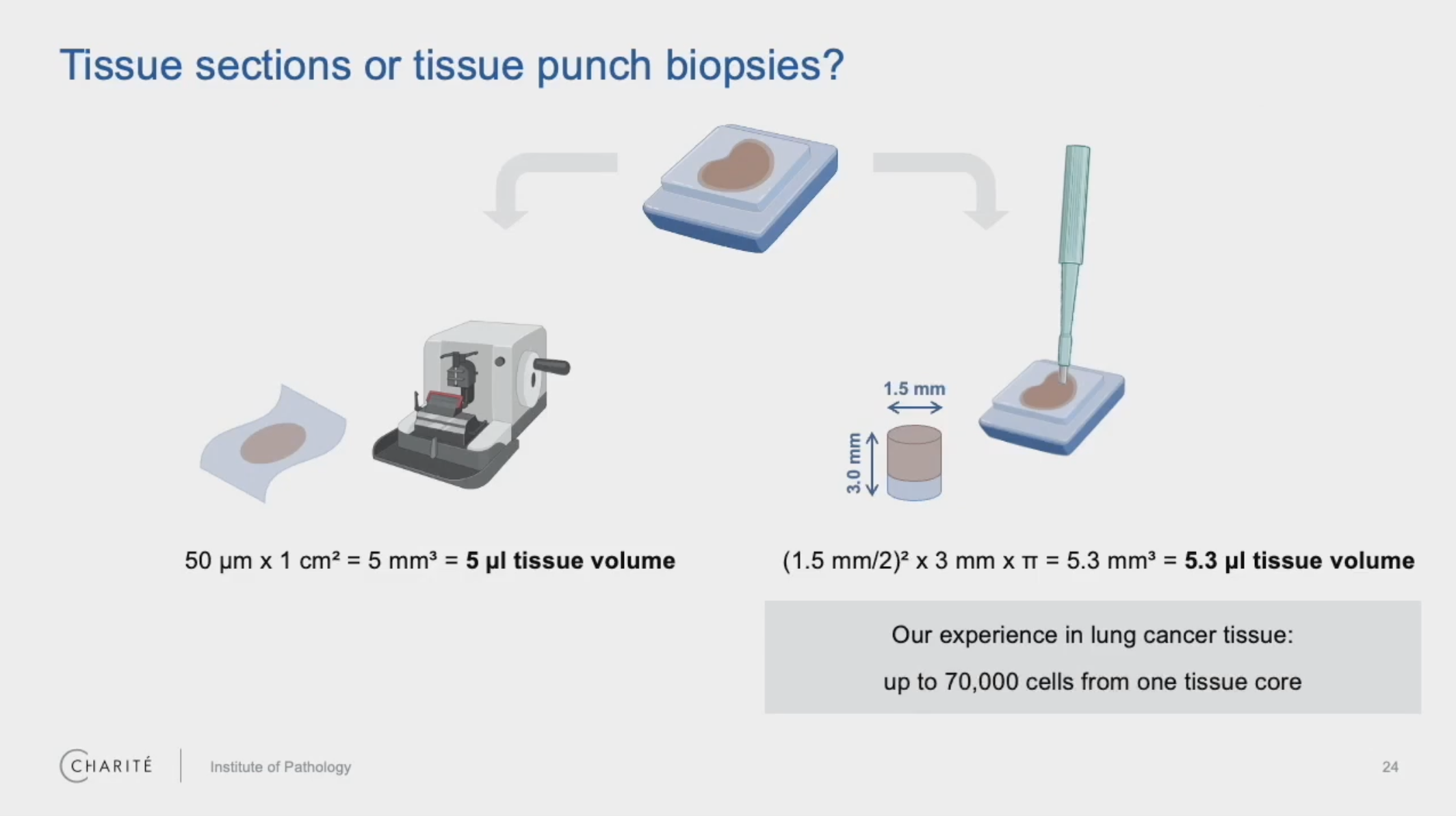
Are there any size limitations? Are microarray cores, for example, too small to analyze?
Bischoff: So, first, this depends on the type of tissue. If you have very dense tissue of high cellularity, then you need, of course, less volume. If you have tissue of less cellularity, you need more tissue volume. In the non-small cell lung cancers, which have higher cellularity, the minimum amount was one tissue core, which I also showed in my talk. So, one core of 1 or 1.5 mm in diameter.
So, one core of a tissue microarray (TMA) might be enough, but you will use the entire core of the TMA. You will not be able to do additional stainings on this FFPE block. This might be the minimum amount of input tissue you will need for your analysis.
*Here, cores refer to tissue punch biopsies, while curls refer to a whole section of tissue, both taken from FFPE blocks.
Data quality and gene expression profiles
Were there any patterns to which types of differentially expressed genes were poorly correlated across the fresh tissue and the FFPE tissue?
Bischoff: Good question. So, we did not see that there is a specific family of genes which is poorly correlated. It rather seemed that lowly expressed genes maybe show a bit less correlation, but highly expressed genes—for example, cell-type markers—showed a very good correlation. So it might be related to the expression strength of the genes.
Are there particular immune cells that are not well represented in the FFPE samples versus fresh frozen? For example, did you see differences in the representation of myeloid cells between the FFPE and fresh frozen samples?
Bischoff: We didn't observe that. Actually, we found that all cell types in the FFPE tissue were also found in the fresh tissue. This was quite comparable. As for the other immune cells, the quantity of cell types and cell subtypes was quite comparable.
You can learn more about single cell profiling of FFPE with Gene Expression Flex by visiting the product page or this recent sample prep blog with a wealth of helpful resources and advice from our scientists. Do you have specific questions on your tissue of interest? Check out our list of tested tissues, provided by our technical support team.
This Q&A has been edited for length and clarity. Watch the full, on-demand webinar here.
