Finding the perfect fit: How to choose the right single cell assay
What does your single cell research look like? Are you exploring drug response and resistance, investigating immune response to vaccination, tracking Alzheimer’s disease pathology, or trying to uncover drivers of cancer development and patient outcomes? Maybe you’re building a comprehensive cell atlas to lay a solid foundation for the exploration of healthy development and disease progression. Scientists have been taking advantage of single cell profiling in these and other fields for over a decade, identifying cell subpopulations, capturing rare cell types, and determining cell state. These information-rich datasets enable explorations into complex biology and help reveal critical insights into the underlying cellular changes at work in health and disease.
While each project prioritizes different research goals, one common question comes to the forefront: how do you best make use of your samples, time, and budget? All of these are precious resources and choosing the best assay for your experiment can go a long way to ensuring your success. So let’s figure out how to help you decide.
In this blog, we’ll cover practical considerations for experimental setup (including species compatibility, sample types, throughput needs, analyte capture, and more) and discuss how different assays can meet those concerns. While we strongly encourage you to read the discussions that follow, first check out the summary table below for a look at the breadth of applications covered by Chromium single cell assays.
Be sure to keep reading for details, with helpful links to more in-depth information and convenient recaps along the way.
Jump to:
- Single cell assays summary table
- Introducing Flex and Universal
- Flex Gene Expression
- Universal 3’ and 5’ Gene Expression
- Comparing Universal 3’ and 5’ gene expression
At a glance: Chromium single cell gene expression assays
Overall, Flex is our recommended single cell gene expression assay due to its high performance, flexibility, and low sequencing costs. For exceptional results in broader species-agnostic applications or immunoprofiling, we offer Universal 3’ and 5’ assays. An easy, side-by-side comparison of assay capabilities is available in the following summary table, and we further explore each of these attributes in the discussion below.
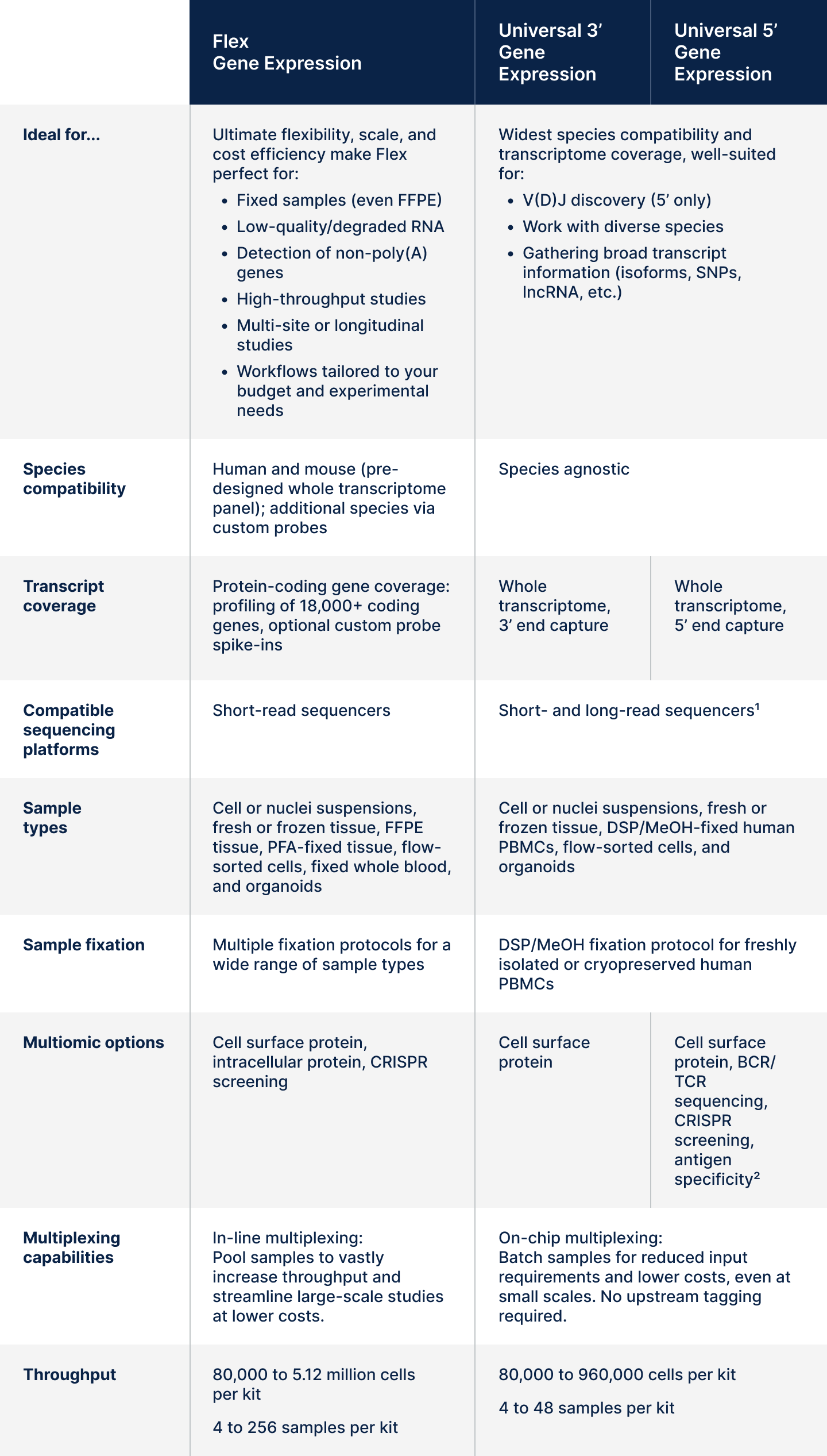
Table 1. Overall summary of assay features and capabilities for Flex, Universal 3’, and Universal 5’ Gene Expression. 1Long-read sequencing for Universal assays is compatible with Oxford Nanopore Technologies' (ONT) PromethION sequencer. Details can be found in this Tech Note. 2Third-party reagents may be used for T- or B-cell antigen specificity experiments in combination with the Universal 5’ assay, as described in this support article.
Already know which assay fits your research? Speak with a Sales Specialist to get started. Still not sure or want to keep exploring? Read on!
Two gene expression assay families: Flex and Universal
Your research has particular parameters that must be met in order to answer key questions. Whether you’re talking about sample collection logistics, RNA quality, scale, or the cellular features you’re trying to capture, this list of requirements informs the method you choose.
Thankfully, there’s a diverse selection of assays to choose from, each with their own set of capabilities. Here, we'll discuss the flagship scRNA-seq assay families on the Chromium platform: Flex and Universal Gene Expression. Both provide highly sensitive transcriptomic data with multiomic options and cover a wide breadth of applications. Chemistry differences between the two lead to different benefits for researchers.
Even with these differences, the overall workflows are similar—and the key to all of them is the ability to efficiently and reliably generate large numbers of GEMs (Gel Beads-in-Emulsions). GEMs are partitions containing micro-reactions in which molecules of interest (including RNA, protein, etc.) from each cell are captured and barcoded. All transcripts from the same cell will share a common barcode, enabling each one to be traced back to its cell of origin.
Barcoded transcripts for hundreds to tens of thousands of cells are then pooled for downstream reactions to create sequencer-compatible libraries. With this solid foundation, the differences in chemistry inside those GEMs are what determine how biological information is captured.
So which assay family is better suited for your scientific inquiries? In short, the unique Flex chemistry makes for an incredibly powerful assay that enables cost savings and experimental flexibility alongside peak performance, and our experts recommend Flex for most single cell applications. For research involving V(D)J discovery or work that requires a species-agnostic approach, our Universal kits have you covered. In the sections below, we’ll expand more on these chemistries and the capabilities each one brings to your bench.
Looking for epigenomic tools? Our Epi assay family enables single cell profiling of chromatin accessibility. Analysis of epigenomic information can reveal areas of active gene transcription, as well as regulatory regions and binding site motifs. While this blog will not go into detail on the Epi kits, the Universal 3’ chemistry discussed below is paired with ATAC-seq in our Epi Multiome kit to achieve direct linkage of gene expression and open chromatin readouts from the same nucleus.
Flex Gene Expression
The Flex Gene Expression assay provides highly sensitive protein-coding gene coverage for human or mouse samples, with options for customizable coverage to address specific experimental needs. Notably, Flex yields high-quality results from samples with low-quality or damaged RNA. This method also enables more flexible workflow options and sample-type compatibility, including fresh, frozen, and fixed samples—even FFPE sections and fixed whole blood. Quick and easy fixation protocols preserve biology while reducing urgency from sample processing and streamlining experimental logistics.
Why might you choose the Flex assay?
This method is preferred for research that requires:
- High sensitivity for protein-coding genes in human or mouse, plus targeted discovery for non-poly(A) genes, low-expressing targets, and custom content
- Robust performance across cells, tissues, and samples, including FFPE tissue and samples with damaged RNA
- Cost efficiency—low costs at large scale, plus high sequencing efficiency for thorough analysis with less sequencing required
- Largest available throughput (over 5 millions cells per kit)
- Easy, flexible workflows for fixation, storage, and batching—ideal for longitudinal or multi-site studies
The Flex whole transcriptome panel covers 18,000+ protein-coding genes from human or mouse, optimized for high performance with three-fold coverage of targets and a small, 50-bp footprint. These features maximize sensitivity, even with degraded RNA. To accommodate additional experimental needs, the pre-designed panel can be supplemented with your own custom probes for transcripts of interest, including other species (e.g., Macaque), pathogen detection (e.g., microbial or viral genes), genes not represented in current probe panels (e.g., lncRNAs), noncoding RNAs, or custom transgenes (e.g., EGFP).
Data from side-by-side comparisons between Flex and Universal 3’ show high correlation between the two assays. Additionally, genes that may be hard to detect with the 3’ assay can be robustly detected with Flex. (Further details on the Flex whole transcriptome probe panel can be found here.)
The Flex assay also provides high sequencing efficiency, enabling better detection of low-abundance transcripts, more reliable profiling with less sequencing, and bigger savings for your sequencing budget. Sequencing depth for any given experiment will depend on sample type and the specific research question. But for analysis of new samples with the Flex assay, our scientists recommend sequencing a minimum of 10,000 read pairs per cell for gene expression and 5,000 read pairs per cell for protein analysis.
This probe-based chemistry has further advantages in expanded sample-type compatibility, unique sample prep options, and highly flexible workflows that put less strain on your schedule, no matter what your sample collection process looks like.
Jump to:
- Single cell assays summary table
- Universal 3’ and 5’ Gene Expression
- Comparing Universal 3’ and 5’ gene expression
Sample prep options
As part of the Flex protocol, samples go through a quick and easy fixation step prior to hybridization with transcriptomic probes, and the resulting fixed samples can be stored without compromising downstream data quality. This means accurate biology is preserved at the point of collection, and you can customize your workflow with multiple, optional stopping points.
What practical impact does this have on your experiment?
Sample fixation enables more flexibility in your schedule—you can process samples in batches, even from multiple sites or on different collection schedules, and then run them all together at a later time. You can save extra portions of your samples in case you need to troubleshoot or perform repeat runs. Fixation can also lend more consistency to sample handling across experiments, thus limiting technical artifacts and avoiding skewed analysis.
Flex is compatible with an incredible range of sample types for human and mouse, including cell and nuclei suspensions, fresh and frozen tissue, FFPE sections, PFA-fixed tissue, flow-sorted cells, organoids, and fixed whole blood. A deep dive into sample types, best practices for storage and transport, QC considerations, optional stopping points, and more can be found here.
Multiomic readouts
Multiomic options for the Flex assay yield information-rich data for more comprehensive views of biology. Flex enables capture of cell surface and/or intracellular proteins alongside gene expression from the same cells for analyses that can lead to finer cell-type characterization and more in-depth understanding of cellular pathways. The assay is also compatible with high-throughput functional genomics screens with the use of custom probes for detection of guide RNAs, letting you directly assess CRISPR-driven perturbations and the resulting gene expression phenotypes.
Flex multiomic options
- Gene expression
- Cell surface protein
- Intracellular protein
- gRNAs for CRISPR screens
To explore all applicable Demonstrated Protocols and Knowledge Base articles for this assay, check out our support pages for Flex Gene Expression.
Throughput options
Flex provides multiple throughput options that let you easily move from small- to large-scale studies. In fact, Flex has our highest available throughput, with capacity to profile millions of cells at less than $0.01 per cell.
Throughput capacity:
- 80,000 to 5.12 million cells per kit
- 4 to 256 samples per kit
Two different workflows are available: standard singleplex and in-line multiplex. In-line multiplexing enables your largest studies, including atlasing, multi-site translational studies, large patient cohort studies, and more. Batch your samples and streamline high-throughput experiments at a fraction of the cost.
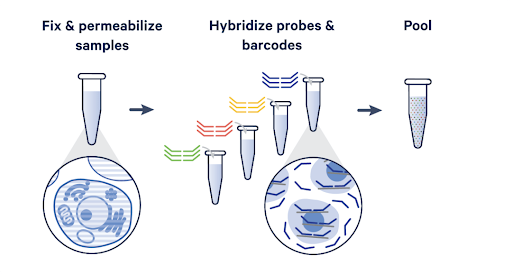
Fixation of cell suspensions and hybridization prior to GEM generation allows for pooling of up to 16 samples per channel, enabling your most ambitious research and bringing convenience to large-scale workflows.
At a glance: Flex assay
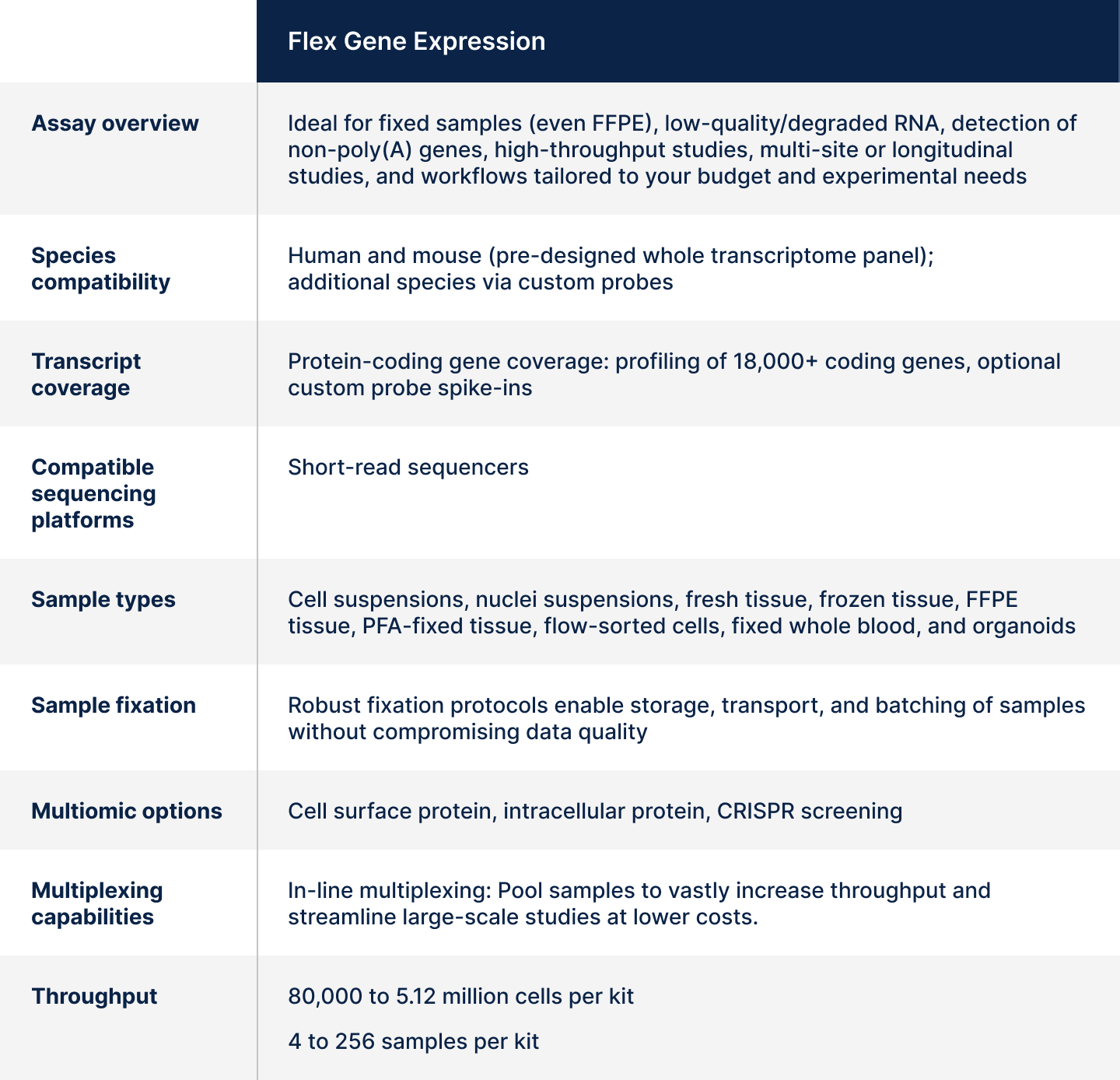
Table 2. Overall summary of assay features and capabilities for Flex Gene Expression.
Universal 3’ and 5’ Gene Expression
As with Flex Gene Expression, Universal 3’ and 5’ Gene Expression assays provide high-performance, reliable single cell profiling for a wide breadth of applications. These assays generate whole transcriptome single cell libraries that represent the 3’ and 5’ ends of the transcript, respectively, and enable capture of a wide range of multiomic readouts for more comprehensive cell-type characterization. These methods are species agnostic and are compatible with cell or nuclei suspensions, fresh or frozen tissue, flow-sorted cells, DSP/methanol-fixed human PBMCs, and organoids.
Why might you choose a Universal assay?
These assays are preferred for research that requires:
- Gathering broad information (isoforms, single nucleotide polymorphisms [SNPs], long non-coding RNAs [lncRNAs], etc.)
- Working with diverse species
- Performing V(D)J discovery
- Long-read sequencing (compatible with Oxford Nanopore Technologies’ PromethION sequencers)
Both 3’ and 5’ assays employ reverse transcription (RT)–based chemistry and use poly-dT primers for RT, but they each capture different ends of the polyadenylated transcript in the final library.
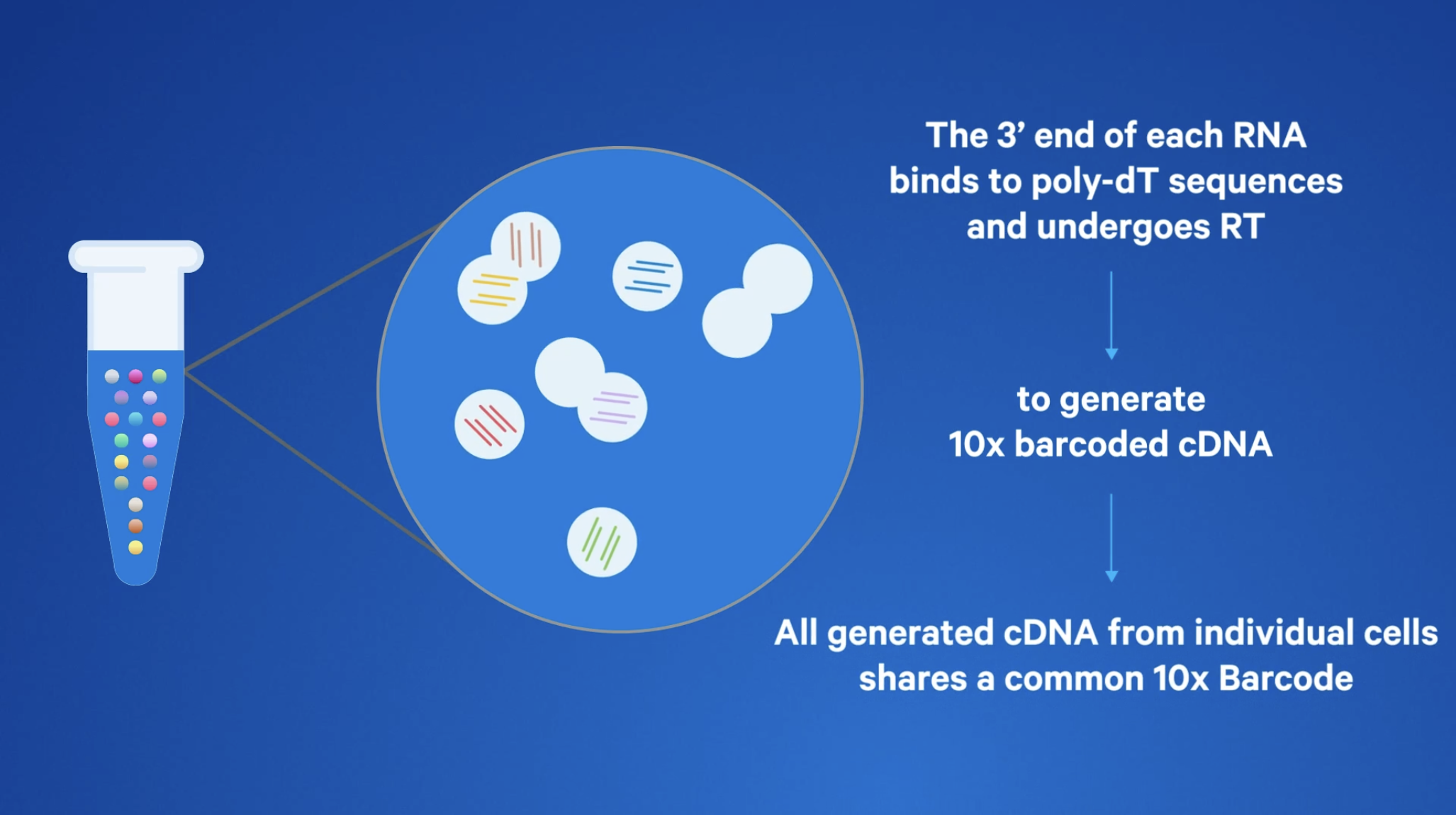
Final libraries generated with Universal assays represent either the 3' or 5’ end of the transcript. One may be more suitable than the other, depending on multiomic capabilities or the capture of information on regulatory mechanisms at the different ends of the transcript.
Jump to:
Comparing and contrasting Universal 3’ and 5’ gene expression
These two assays have many similarities, from throughput capacity to sample compatibility. But one key difference—capture of the 3’ or 5’ end of the transcript—impacts the types of information that can be profiled beyond gene expression.
Exploring regulatory mechanisms
3’ assays are commonly used for gene expression profiling and identification of cell types and subtypes. Capture of the 3’ end of the polyadenylated transcript also enables researchers to acquire information on alternative polyadenylation (APA). Studies have shown APA to be widespread—observed in ~70% of mammalian genes and ~50% of human genes (1,2,3). While scientists find tight control on APA during healthy development, dysregulation is seen in disease, with noted links to cancer progression (2,3).
Capturing the 5’ end of the transcript lets you study transcription start sites (TSS) and alternative splicing. For example, the 5’ assay can allow for exploration of differential TSS usage in individual cells. This variation accounts for altered lengths of 5’ UTRs, and research has shown downstream effects on transcript stability and transcript diversity (4,5). TSS regulation has also been linked with cell-fate transition and patient prognosis in some cancers (5).
Multiomic readouts and additional experimental options
The 3' and 5' kits also offer different sets of multiomic capabilities. If we look at the 3’ assay, the addition of cell surface protein readouts can enable further refinement of cell subpopulations or provide information on cell state. The 5’ kit also provides cell surface protein data and has additional options for multiomic readouts. Simultaneous capture of guide RNAs (gRNAs) and the resulting perturbations enables high-throughput CRISPR screens. Notably, key readouts are available for immune profiling studies, with full-length V(D)J sequences for immune cell characterization, plus options that allow for antigen specificity studies.
3’ multiomic options
- 3’ gene expression
- Cell surface protein
5’ multiomic options
- 5’ gene expression
- Cell surface protein
- gRNAs for CRISPR screens
- Paired full-length BCR/TCR genes
- Antigen specificity studies
For epigenomic applications, check out our Epi ATAC kit or see how Universal 3’ chemistry is paired with ATAC-seq in our Epi Multiome kit to achieve simultaneous capture of gene expression and chromatin accessibility from the same individual cells.
To explore all applicable Demonstrated Protocols and Knowledge Base articles for Universal assays, check out our support pages for Universal 3’ Gene Expression and Universal 5’ Gene Expression.
Throughput options
As with the Flex assay, Universal 3‘ and 5’ Gene Expression assays provide flexible throughput options that let you easily scale from small pilot studies to more ambitious experiments.
Throughput capacity:
- 80,000 to 960,000 cells per kit
- 4 to 48 samples per kit
For more experienced single cell researchers who want to go beyond this capacity, internal testing demonstrates that our Universal assays are compatible with Cell Hashing, enabling even higher throughputs. Details on how to perform Cell Hashing for the 3’ and 5’ kits can be found here and here, respectively.
Two different standard workflows are available for the Universal assay family: singleplex and on-chip multiplex. On-chip multiplexing is a simple and reliable workflow that reduces reagent volumes and enables more cost-effective studies, even at small scales. With lower costs per sample, you can add biological and technical replicates in a cost-effective manner. This method also eases technical restrictions with lower cell inputs (up to 5,000 cells per sample for each run) for multiplexing of limited samples like stem cells or flow-sorted cells for rare disease studies.
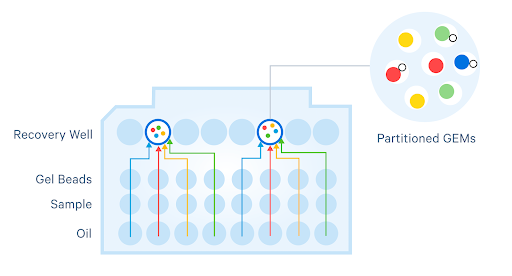
The protocol is easy and streamlined—cells from 4 samples are co-partitioned and GEMs from all 4 are collected in the same recovery well, no need to tag samples upstream of chip loading. Aside from not introducing any extra steps, the lack of upstream tagging means this protocol is compatible with diverse species.
At a glance: Universal 3’ and 5’ assays
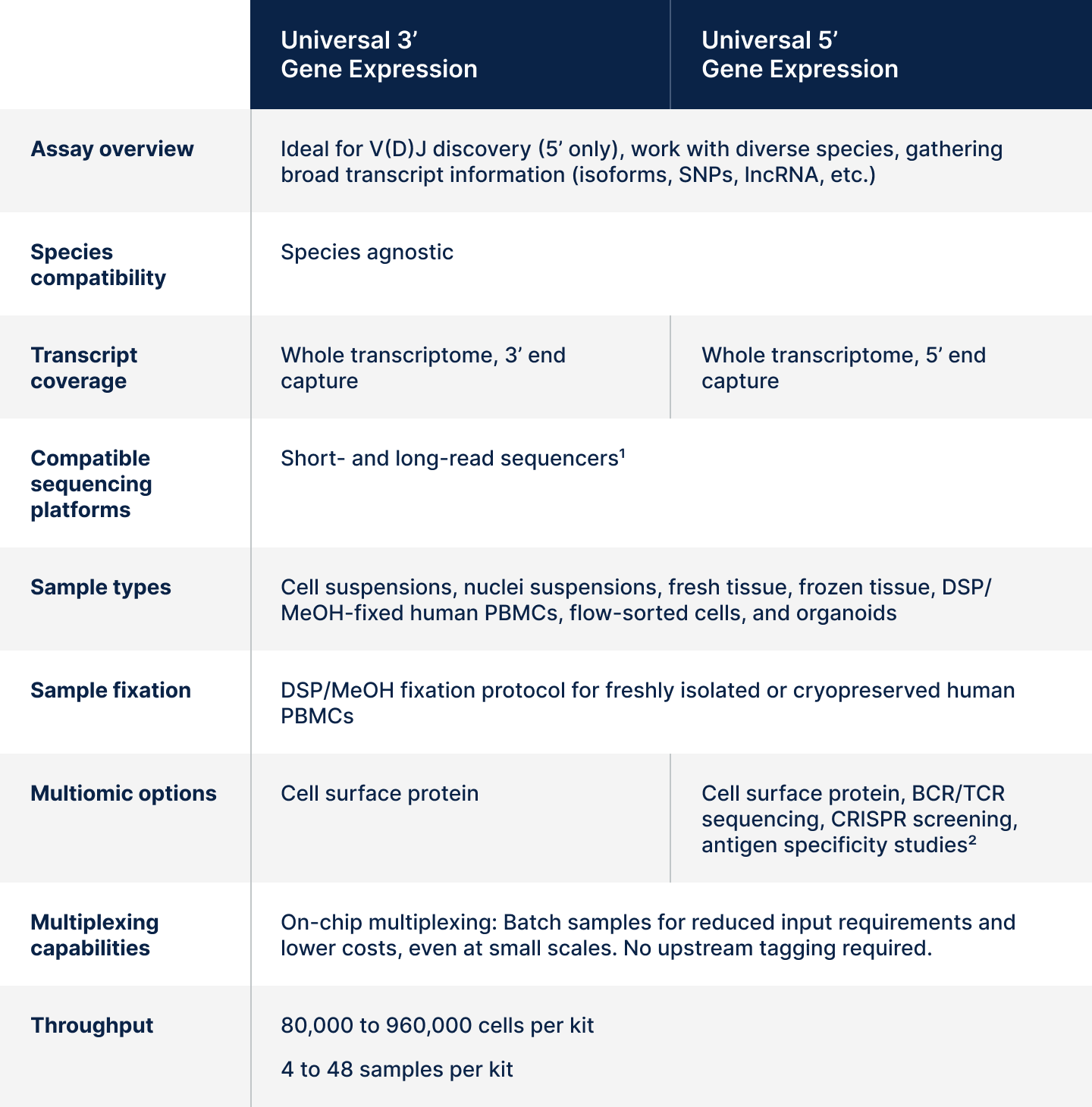
Table 3. Overall summary of assay features and capabilities for Universal 3’ and 5’ Gene Expression. 1Long-read sequencing for Universal assays is compatible with Oxford Nanopore Technologies' (ONT) PromethION sequencer. Details can be found in this Tech Note. 2Third-party reagents may be used for T or B cell antigen specificity experiments in combination with the Universal 5’ assay, as described in this support article.
Recapping and next steps
We’ve seen that single cell solutions cover a wide range of experimental needs, but the key to your success is letting the science lead the way. With that in mind, we invite you to take another look at the summary table and see how each assay fits into your research goals, revisiting the key decision points, from species and sample type to throughput and multiomic analysis.
Visit the single cell applications page to continue exploring and choose your assay. If you still aren’t sure which one would be best for your research, we’re happy to help. Contact us to speak with a specialist.
For a more in-depth look at Chromium technology, download the tech brochure.
References:
- Fansler M, et al. Comprehensive annotation of 3′UTRs from primary cells and their quantification from scRNA-seq data. bioRxiv (2023). doi: 10.1101/2021.11.22.469635
- Agarwal V, et al. The landscape of alternative polyadenylation in single cells of the developing mouse embryo. Nat Commun 12: 5101 (2021). doi: 10.1038/s41467-021-25388-8
- Kandhari N, et al. The detection and bioinformatic analysis of alternative 3' UTR isoforms as potential cancer biomarkers. Int J Mol Sci 22(10): 5322 (2021). doi: 10.3390/ijms22105322
- Policastro R, et al. Flexible analysis of TSS mapping data and detection of TSS shifts with TSRexploreR. NAR Genom Bioinform 3(2): lqab051 (2021). doi: 10.1093/nargab/lqab051
- Hou R, et al. CamoTSS: analysis of alternative transcription start sites for cellular phenotypes and regulatory patterns from 5' scRNA-seq data. Nat Commun 14: 7240 (2023). doi: 10.1038/s41467-023-42636-1
