Genetics lab at Stanford releases a new data resource for benchmarking genome analysis pipelines
The extensive collection of Illumina sequencing datasets, described by Bo Zhou and his co-authors (Alexander Urban Lab, Depts. of Psychiatry and Genetics) in a recent bioRxiv preprint, are derived from J. Craig Venter’s genome. The Venter human reference genome assembly (HuRef) has the distinction of being the only human genome for which a diploid Sanger-sequencing-based assembly was generated (Levy et al. 2007). It has been used extensively to study genome variation, and there are now more than 76 catalogs of validated variants available (Pang et al. 2010; Mu et al. 2015). Thus, the HuRef assembly serves as a validated comparator for those seeking to test and tune new genome analysis tools.
Zhou and his co-authors prepared and sequenced multiple paired-end and mate-pair libraries, as well as a single Chromium Linked-Read library. The Linked-Read library was sequenced to 133x physical coverage (2x150bp reads, HiSeqX with single indexing) and used to perform haplotype phasing and variant calling with the Long Ranger (V2.1.3) analysis pipelines. The phasing results and variant calling performance metrics are shown below (Table).
TABLE: Metrics for Linked-Read sequencing and phasing of the Venter genome*.
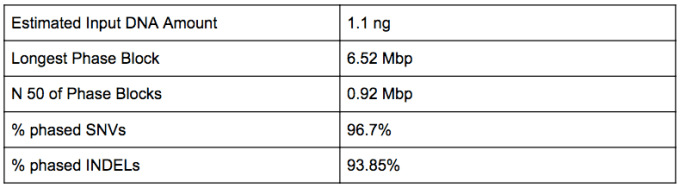
All of the whole genome sequence datasets produced in this study have been deposited in the NCBI sequence read archive. Additionally, the original vcf file of phased variants from the Linked-Read analysis is available through dbSNP. Tool developers can now draw on this rich collection of data provided by the team at Stanford to test their algorithms that use Illumina short-read data and directly benchmark results against the high-quality, validated HuRef assembly.
Learn More:
-
Read the BioRxiv Preprint: Extensive and deep sequencing of the Venter/HuRef genome for developing and benchmarking genome analysis tools
-
Find literature and watch videos about Linked-Read Sequencing on the Chromium Genome Sequencing Solution page
