High-Definition Immunology: Paired Immune Repertoire Profiling at Single-Cell Resolution
The adaptive immune response, though critical in any understanding of human health, has proven challenging to study. Immune response is largely governed by two factors—lymphocyte diversity and antigen specificity. Healthy individuals typically have hugely diverse populations of T cells, each with an antigen-specific T-cell Receptor (TCR) formed from diverse alpha and beta chain genes. This TCR diversity is generated through V(D)J Recombination, the somatic recombination of Variable (V), Diversity (D), and Joining (J) gene sequences. This process creates sequence variability at the V/D and D/J recombination junctions, providing T cells with a layer of genetic diversity that does not exist in other cell types and can be measured only with a system that can profile the exact V, D, and J sequences for both alpha and beta genes within each individual cell.
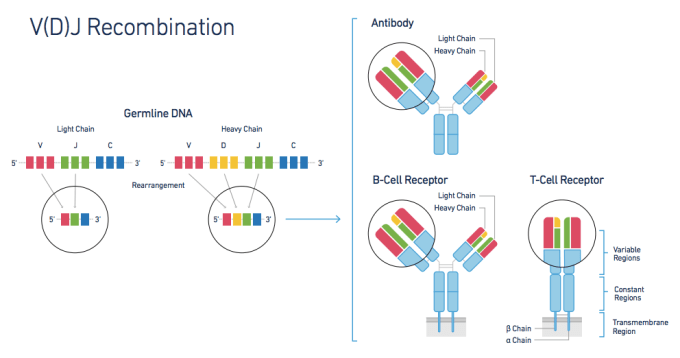
In a recent article in GEN, Brian Fritz, Ph.D. describes how the Chromium™ Single Cell V(D)J Solution solves this problem and reveals true T-cell diversity. The V(D)J system combines microfluidics and 5' molecular barcoding to assemble full-length V(D)J sequences on a cell-by-cell basis. Then, the accompanying bioinformatics software, the Cell Ranger™ pipeline, profiles and groups the V(D)J sequences to build clonotypes, groups of T cells sharing the same TCRs. To validate the system's performance, 10x researchers performed several experiments, profiling a variety of samples containing T cells.
In the article, Fritz describes one of the experiments, wherein T cells exposed to the Epstein-Barr Virus (EBV) were encapsulated and sequenced with the Chromium System and then sorted into clonotypes. They found that 55% of the sequenced T cells shared one major alpha and beta chain, 9% shared a second, unrelated clonotype, and 4% had two related clonotypes that shared a common beta chain. Limit of detection (LOD) experiments were then performed, mixing EBV-specific T cells into samples of PBMCs from a healthy donor at a 1:99 ratio. They expected that the four discovered clonotypes would still be observable at the appropriate levels. Accordingly, the clonotypes were detected at 0.4%, 0.3% and 0.05%, respectively.
Profiling paired V(D)J transcripts from hundreds to millions of lymphocytes on a cell-by-cell basis, the Chromium Single Cell V(D)J system is a complete, scalable tool that has the potential to accelerate progress in immune therapy development. It could greatly impact areas such as vaccine development, clonal immune-cell dynamics, immune responses to checkpoint blockades, and the development of recombinant antibodies and engineered T cells used in immunotherapies for cancer and other diseases.
Additional Resources
- Learn more about the Chromium Single Cell V(D)J Solution
- See the Chromium Single Cell V(D)J Software Suite for assembling, analyzing and visualizing V(D)J sequences and clonotypes
- Download datasets
